Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
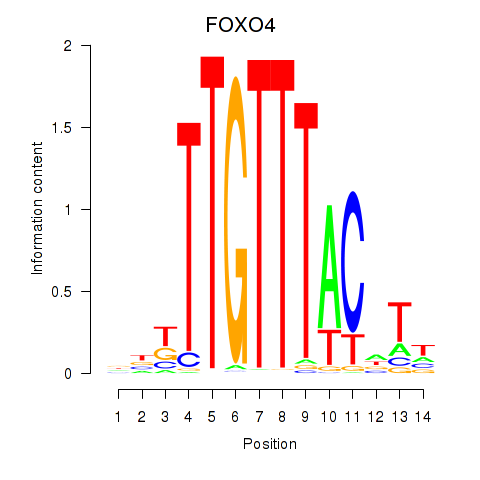
Results for FOXO4
Z-value: 0.50
Transcription factors associated with FOXO4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXO4
|
ENSG00000184481.12 | forkhead box O4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXO4 | hg19_v2_chrX_+_70316005_70316047 | 0.95 | 4.9e-02 | Click! |
Activity profile of FOXO4 motif
Sorted Z-values of FOXO4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_12679171 | 0.42 |
ENST00000606790.1
|
RP11-474O21.5
|
RP11-474O21.5 |
| chr3_-_129375556 | 0.37 |
ENST00000510323.1
|
TMCC1
|
transmembrane and coiled-coil domain family 1 |
| chr10_-_90751038 | 0.37 |
ENST00000458159.1
ENST00000415557.1 ENST00000458208.1 |
ACTA2
|
actin, alpha 2, smooth muscle, aorta |
| chr9_-_88896977 | 0.29 |
ENST00000311534.6
|
ISCA1
|
iron-sulfur cluster assembly 1 |
| chr14_+_50234827 | 0.28 |
ENST00000554589.1
ENST00000557247.1 |
KLHDC2
|
kelch domain containing 2 |
| chr3_+_187461442 | 0.26 |
ENST00000450760.1
|
RP11-211G3.2
|
RP11-211G3.2 |
| chr9_+_67977438 | 0.25 |
ENST00000456982.1
|
RP11-195B21.3
|
Protein LOC644249 |
| chr3_+_148447887 | 0.23 |
ENST00000475347.1
ENST00000474935.1 ENST00000461609.1 |
AGTR1
|
angiotensin II receptor, type 1 |
| chr15_-_55657428 | 0.22 |
ENST00000568543.1
|
CCPG1
|
cell cycle progression 1 |
| chr9_+_42671887 | 0.22 |
ENST00000456520.1
ENST00000377391.3 |
CBWD7
|
COBW domain containing 7 |
| chr4_-_155511887 | 0.22 |
ENST00000302053.3
ENST00000403106.3 |
FGA
|
fibrinogen alpha chain |
| chr9_-_86432547 | 0.22 |
ENST00000376365.3
ENST00000376371.2 |
GKAP1
|
G kinase anchoring protein 1 |
| chr1_-_241803649 | 0.20 |
ENST00000366554.2
|
OPN3
|
opsin 3 |
| chr5_+_162932554 | 0.20 |
ENST00000321757.6
ENST00000421814.2 ENST00000518095.1 |
MAT2B
|
methionine adenosyltransferase II, beta |
| chr8_-_101348408 | 0.19 |
ENST00000519527.1
ENST00000522369.1 |
RNF19A
|
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
| chr11_+_77532233 | 0.19 |
ENST00000525409.1
|
AAMDC
|
adipogenesis associated, Mth938 domain containing |
| chr7_-_95225768 | 0.19 |
ENST00000005178.5
|
PDK4
|
pyruvate dehydrogenase kinase, isozyme 4 |
| chr1_-_241803679 | 0.19 |
ENST00000331838.5
|
OPN3
|
opsin 3 |
| chr12_+_121416437 | 0.19 |
ENST00000402929.1
ENST00000535955.1 ENST00000538626.1 ENST00000543427.1 |
HNF1A
|
HNF1 homeobox A |
| chr4_-_152149033 | 0.17 |
ENST00000514152.1
|
SH3D19
|
SH3 domain containing 19 |
| chr3_+_160394940 | 0.17 |
ENST00000320767.2
|
ARL14
|
ADP-ribosylation factor-like 14 |
| chr5_-_125930877 | 0.17 |
ENST00000510111.2
ENST00000509270.1 |
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1 |
| chr3_-_28389922 | 0.17 |
ENST00000415852.1
|
AZI2
|
5-azacytidine induced 2 |
| chr4_+_124571409 | 0.17 |
ENST00000514823.1
ENST00000511919.1 ENST00000508111.1 |
RP11-93L9.1
|
long intergenic non-protein coding RNA 1091 |
| chr3_+_101292939 | 0.17 |
ENST00000265260.3
ENST00000469941.1 ENST00000296024.5 |
PCNP
|
PEST proteolytic signal containing nuclear protein |
| chr17_-_1418972 | 0.17 |
ENST00000571274.1
|
INPP5K
|
inositol polyphosphate-5-phosphatase K |
| chr4_+_76871883 | 0.17 |
ENST00000599764.1
|
AC110615.1
|
Uncharacterized protein |
| chr5_-_125930929 | 0.16 |
ENST00000553117.1
ENST00000447989.2 ENST00000409134.3 |
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1 |
| chr16_-_15736881 | 0.16 |
ENST00000540441.2
|
KIAA0430
|
KIAA0430 |
| chr16_+_84801852 | 0.16 |
ENST00000569925.1
ENST00000567526.1 |
USP10
|
ubiquitin specific peptidase 10 |
| chr2_+_196521903 | 0.16 |
ENST00000541054.1
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr2_-_128615517 | 0.16 |
ENST00000409698.1
|
POLR2D
|
polymerase (RNA) II (DNA directed) polypeptide D |
| chr10_-_25241499 | 0.15 |
ENST00000376378.1
ENST00000376376.3 ENST00000320152.6 |
PRTFDC1
|
phosphoribosyl transferase domain containing 1 |
| chr4_-_14889791 | 0.15 |
ENST00000509654.1
ENST00000515031.1 ENST00000505089.2 |
LINC00504
|
long intergenic non-protein coding RNA 504 |
| chr1_+_63989004 | 0.15 |
ENST00000371088.4
|
EFCAB7
|
EF-hand calcium binding domain 7 |
| chr5_-_42811986 | 0.15 |
ENST00000511224.1
ENST00000507920.1 ENST00000510965.1 |
SEPP1
|
selenoprotein P, plasma, 1 |
| chr4_+_24661479 | 0.15 |
ENST00000569621.1
|
RP11-496D24.2
|
RP11-496D24.2 |
| chr14_-_64108125 | 0.15 |
ENST00000267522.3
|
WDR89
|
WD repeat domain 89 |
| chr12_+_96588368 | 0.15 |
ENST00000547860.1
|
ELK3
|
ELK3, ETS-domain protein (SRF accessory protein 2) |
| chr4_+_172734548 | 0.15 |
ENST00000506823.1
|
GALNTL6
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 6 |
| chr14_-_102605983 | 0.15 |
ENST00000334701.7
|
HSP90AA1
|
heat shock protein 90kDa alpha (cytosolic), class A member 1 |
| chr7_+_134551583 | 0.15 |
ENST00000435928.1
|
CALD1
|
caldesmon 1 |
| chr15_-_31283798 | 0.14 |
ENST00000435680.1
ENST00000425768.1 |
MTMR10
|
myotubularin related protein 10 |
| chr15_-_52970820 | 0.14 |
ENST00000261844.7
ENST00000399202.4 ENST00000562135.1 |
FAM214A
|
family with sequence similarity 214, member A |
| chr5_-_10308125 | 0.14 |
ENST00000296658.3
|
CMBL
|
carboxymethylenebutenolidase homolog (Pseudomonas) |
| chr2_+_109204743 | 0.14 |
ENST00000332345.6
|
LIMS1
|
LIM and senescent cell antigen-like domains 1 |
| chr18_+_72922710 | 0.14 |
ENST00000322038.5
|
TSHZ1
|
teashirt zinc finger homeobox 1 |
| chr12_-_12491608 | 0.13 |
ENST00000545735.1
|
MANSC1
|
MANSC domain containing 1 |
| chr14_+_38065052 | 0.13 |
ENST00000556845.1
|
TTC6
|
tetratricopeptide repeat domain 6 |
| chr4_-_139163491 | 0.13 |
ENST00000280612.5
|
SLC7A11
|
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
| chr3_+_130569429 | 0.13 |
ENST00000505330.1
ENST00000504381.1 ENST00000507488.2 ENST00000393221.4 |
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr9_+_5510558 | 0.13 |
ENST00000397747.3
|
PDCD1LG2
|
programmed cell death 1 ligand 2 |
| chr11_-_77531752 | 0.13 |
ENST00000440064.2
ENST00000528095.1 |
RSF1
|
remodeling and spacing factor 1 |
| chr17_-_46806540 | 0.12 |
ENST00000290295.7
|
HOXB13
|
homeobox B13 |
| chr9_-_134585221 | 0.12 |
ENST00000372190.3
ENST00000427994.1 |
RAPGEF1
|
Rap guanine nucleotide exchange factor (GEF) 1 |
| chr3_-_49066811 | 0.12 |
ENST00000442157.1
ENST00000326739.4 |
IMPDH2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2 |
| chr6_-_13621126 | 0.12 |
ENST00000600057.1
|
AL441883.1
|
Uncharacterized protein |
| chr6_+_108882069 | 0.12 |
ENST00000406360.1
|
FOXO3
|
forkhead box O3 |
| chr2_+_196521845 | 0.12 |
ENST00000359634.5
ENST00000412905.1 |
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr9_-_136933615 | 0.12 |
ENST00000371834.2
|
BRD3
|
bromodomain containing 3 |
| chr7_-_105332084 | 0.12 |
ENST00000472195.1
|
ATXN7L1
|
ataxin 7-like 1 |
| chr2_+_58134756 | 0.12 |
ENST00000435505.2
ENST00000417641.2 |
VRK2
|
vaccinia related kinase 2 |
| chr7_-_95064264 | 0.12 |
ENST00000536183.1
ENST00000433091.2 ENST00000222572.3 |
PON2
|
paraoxonase 2 |
| chr9_+_5510492 | 0.12 |
ENST00000397745.2
|
PDCD1LG2
|
programmed cell death 1 ligand 2 |
| chr5_+_137673200 | 0.12 |
ENST00000434981.2
|
FAM53C
|
family with sequence similarity 53, member C |
| chr2_+_191513959 | 0.12 |
ENST00000337386.5
ENST00000357215.5 |
NAB1
|
NGFI-A binding protein 1 (EGR1 binding protein 1) |
| chr1_+_153940741 | 0.12 |
ENST00000431292.1
|
CREB3L4
|
cAMP responsive element binding protein 3-like 4 |
| chr1_+_28844778 | 0.12 |
ENST00000411533.1
|
RCC1
|
regulator of chromosome condensation 1 |
| chr5_+_156712372 | 0.12 |
ENST00000541131.1
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chr5_+_137673945 | 0.11 |
ENST00000513056.1
ENST00000511276.1 |
FAM53C
|
family with sequence similarity 53, member C |
| chr14_+_56127989 | 0.11 |
ENST00000555573.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr7_+_106810165 | 0.11 |
ENST00000468401.1
ENST00000497535.1 ENST00000485846.1 |
HBP1
|
HMG-box transcription factor 1 |
| chr16_-_79804394 | 0.11 |
ENST00000567993.1
|
RP11-345M22.2
|
RP11-345M22.2 |
| chr1_+_153940346 | 0.11 |
ENST00000405694.3
ENST00000449724.1 ENST00000368607.3 ENST00000271889.4 |
CREB3L4
|
cAMP responsive element binding protein 3-like 4 |
| chr10_+_111985713 | 0.11 |
ENST00000239007.7
|
MXI1
|
MAX interactor 1, dimerization protein |
| chr4_-_100242549 | 0.11 |
ENST00000305046.8
ENST00000394887.3 |
ADH1B
|
alcohol dehydrogenase 1B (class I), beta polypeptide |
| chr11_-_85780853 | 0.11 |
ENST00000531930.1
ENST00000528398.1 |
PICALM
|
phosphatidylinositol binding clathrin assembly protein |
| chr7_+_12250886 | 0.11 |
ENST00000444443.1
ENST00000396667.3 |
TMEM106B
|
transmembrane protein 106B |
| chr17_+_8924837 | 0.11 |
ENST00000173229.2
|
NTN1
|
netrin 1 |
| chr2_-_128615681 | 0.11 |
ENST00000409955.1
ENST00000272645.4 |
POLR2D
|
polymerase (RNA) II (DNA directed) polypeptide D |
| chr5_+_79703823 | 0.10 |
ENST00000338008.5
ENST00000510158.1 ENST00000505560.1 |
ZFYVE16
|
zinc finger, FYVE domain containing 16 |
| chr12_-_31479045 | 0.10 |
ENST00000539409.1
ENST00000395766.1 |
FAM60A
|
family with sequence similarity 60, member A |
| chr18_-_53735601 | 0.10 |
ENST00000589754.1
|
CTD-2008L17.2
|
CTD-2008L17.2 |
| chr8_+_101349823 | 0.10 |
ENST00000519566.1
|
KB-1991G8.1
|
KB-1991G8.1 |
| chr17_+_44668035 | 0.10 |
ENST00000398238.4
ENST00000225282.8 |
NSF
|
N-ethylmaleimide-sensitive factor |
| chr10_-_118032697 | 0.10 |
ENST00000439649.3
|
GFRA1
|
GDNF family receptor alpha 1 |
| chr1_+_81106951 | 0.10 |
ENST00000443565.1
|
RP5-887A10.1
|
RP5-887A10.1 |
| chr14_-_55878538 | 0.10 |
ENST00000247178.5
|
ATG14
|
autophagy related 14 |
| chr13_+_98086445 | 0.10 |
ENST00000245304.4
|
RAP2A
|
RAP2A, member of RAS oncogene family |
| chr12_+_54378923 | 0.10 |
ENST00000303460.4
|
HOXC10
|
homeobox C10 |
| chr1_-_207095324 | 0.10 |
ENST00000530505.1
ENST00000367091.3 ENST00000442471.2 |
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr2_-_148779106 | 0.10 |
ENST00000416719.1
ENST00000264169.2 |
ORC4
|
origin recognition complex, subunit 4 |
| chr1_+_97188188 | 0.10 |
ENST00000541987.1
|
PTBP2
|
polypyrimidine tract binding protein 2 |
| chr5_-_68339648 | 0.09 |
ENST00000479830.2
|
CTC-498J12.1
|
CTC-498J12.1 |
| chr8_-_42698292 | 0.09 |
ENST00000529779.1
|
THAP1
|
THAP domain containing, apoptosis associated protein 1 |
| chr3_-_28390581 | 0.09 |
ENST00000479665.1
|
AZI2
|
5-azacytidine induced 2 |
| chr14_+_96968802 | 0.09 |
ENST00000556619.1
ENST00000392990.2 |
PAPOLA
|
poly(A) polymerase alpha |
| chr8_+_74903580 | 0.09 |
ENST00000284818.2
ENST00000518893.1 |
LY96
|
lymphocyte antigen 96 |
| chr10_-_103578162 | 0.09 |
ENST00000361464.3
ENST00000357797.5 ENST00000370094.3 |
MGEA5
|
meningioma expressed antigen 5 (hyaluronidase) |
| chrX_-_20236970 | 0.09 |
ENST00000379548.4
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| chr3_+_124223586 | 0.09 |
ENST00000393496.1
|
KALRN
|
kalirin, RhoGEF kinase |
| chr14_-_75518129 | 0.09 |
ENST00000556257.1
ENST00000557648.1 ENST00000553263.1 ENST00000355774.2 ENST00000380968.2 ENST00000238662.7 |
MLH3
|
mutL homolog 3 |
| chr1_-_39395165 | 0.09 |
ENST00000372985.3
|
RHBDL2
|
rhomboid, veinlet-like 2 (Drosophila) |
| chr14_+_102276192 | 0.09 |
ENST00000557714.1
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr4_+_184365744 | 0.09 |
ENST00000504169.1
ENST00000302350.4 |
CDKN2AIP
|
CDKN2A interacting protein |
| chr13_+_73629107 | 0.09 |
ENST00000539231.1
|
KLF5
|
Kruppel-like factor 5 (intestinal) |
| chr2_-_113012592 | 0.09 |
ENST00000272570.5
ENST00000409573.2 |
ZC3H8
|
zinc finger CCCH-type containing 8 |
| chr9_-_88897426 | 0.09 |
ENST00000375991.4
ENST00000326094.4 |
ISCA1
|
iron-sulfur cluster assembly 1 |
| chr3_+_130569592 | 0.08 |
ENST00000533801.2
|
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr1_+_145525015 | 0.08 |
ENST00000539363.1
ENST00000538811.1 |
ITGA10
|
integrin, alpha 10 |
| chr7_+_12250943 | 0.08 |
ENST00000442107.1
|
TMEM106B
|
transmembrane protein 106B |
| chr3_+_99536663 | 0.08 |
ENST00000421999.2
ENST00000463526.1 |
CMSS1
|
cms1 ribosomal small subunit homolog (yeast) |
| chr2_+_65283506 | 0.08 |
ENST00000377990.2
|
CEP68
|
centrosomal protein 68kDa |
| chr11_-_107328527 | 0.08 |
ENST00000282251.5
ENST00000433523.1 |
CWF19L2
|
CWF19-like 2, cell cycle control (S. pombe) |
| chr15_+_59397275 | 0.08 |
ENST00000288207.2
|
CCNB2
|
cyclin B2 |
| chr4_-_111120334 | 0.08 |
ENST00000503885.1
|
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr1_-_153514241 | 0.08 |
ENST00000368718.1
ENST00000359215.1 |
S100A5
|
S100 calcium binding protein A5 |
| chr8_+_19171128 | 0.08 |
ENST00000265807.3
|
SH2D4A
|
SH2 domain containing 4A |
| chr1_+_153940713 | 0.08 |
ENST00000368601.1
ENST00000368603.1 ENST00000368600.3 |
CREB3L4
|
cAMP responsive element binding protein 3-like 4 |
| chr4_+_124320665 | 0.08 |
ENST00000394339.2
|
SPRY1
|
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
| chrX_-_38080077 | 0.08 |
ENST00000378533.3
ENST00000544439.1 ENST00000432886.2 ENST00000538295.1 |
SRPX
|
sushi-repeat containing protein, X-linked |
| chr1_+_150229554 | 0.08 |
ENST00000369111.4
|
CA14
|
carbonic anhydrase XIV |
| chr12_+_118454500 | 0.08 |
ENST00000537315.1
ENST00000229043.3 ENST00000484086.2 ENST00000420967.1 ENST00000454402.2 ENST00000392542.2 ENST00000535092.1 |
RFC5
|
replication factor C (activator 1) 5, 36.5kDa |
| chr17_-_30185946 | 0.08 |
ENST00000579741.1
|
COPRS
|
coordinator of PRMT5, differentiation stimulator |
| chrX_+_9431324 | 0.08 |
ENST00000407597.2
ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X
|
transducin (beta)-like 1X-linked |
| chr4_-_70626430 | 0.08 |
ENST00000310613.3
|
SULT1B1
|
sulfotransferase family, cytosolic, 1B, member 1 |
| chr4_-_111120132 | 0.08 |
ENST00000506625.1
|
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr13_-_99630233 | 0.08 |
ENST00000376460.1
ENST00000442173.1 |
DOCK9
|
dedicator of cytokinesis 9 |
| chr5_-_135701164 | 0.07 |
ENST00000355180.3
ENST00000426057.2 ENST00000513104.1 |
TRPC7
|
transient receptor potential cation channel, subfamily C, member 7 |
| chr1_+_200011711 | 0.07 |
ENST00000544748.1
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr16_+_28858004 | 0.07 |
ENST00000322610.8
|
SH2B1
|
SH2B adaptor protein 1 |
| chr4_+_95174445 | 0.07 |
ENST00000509418.1
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr8_+_93895865 | 0.07 |
ENST00000391681.1
|
AC117834.1
|
AC117834.1 |
| chr1_-_227505826 | 0.07 |
ENST00000334218.5
ENST00000366766.2 ENST00000366764.2 |
CDC42BPA
|
CDC42 binding protein kinase alpha (DMPK-like) |
| chr4_-_21950356 | 0.07 |
ENST00000447367.2
ENST00000382152.2 |
KCNIP4
|
Kv channel interacting protein 4 |
| chr4_-_102268628 | 0.07 |
ENST00000323055.6
ENST00000512215.1 ENST00000394854.3 |
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr9_-_39288092 | 0.07 |
ENST00000323947.7
ENST00000297668.6 ENST00000377656.2 ENST00000377659.1 |
CNTNAP3
|
contactin associated protein-like 3 |
| chr16_+_53469525 | 0.07 |
ENST00000544405.2
|
RBL2
|
retinoblastoma-like 2 (p130) |
| chr20_+_44035847 | 0.07 |
ENST00000372712.2
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr3_+_101504200 | 0.07 |
ENST00000422132.1
|
NXPE3
|
neurexophilin and PC-esterase domain family, member 3 |
| chr11_-_46639436 | 0.07 |
ENST00000532281.1
|
HARBI1
|
harbinger transposase derived 1 |
| chr1_+_174670143 | 0.07 |
ENST00000367687.1
ENST00000347255.2 |
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr8_-_40755333 | 0.07 |
ENST00000297737.6
ENST00000315769.7 |
ZMAT4
|
zinc finger, matrin-type 4 |
| chr6_-_105627735 | 0.07 |
ENST00000254765.3
|
POPDC3
|
popeye domain containing 3 |
| chr15_-_52971544 | 0.07 |
ENST00000566768.1
ENST00000561543.1 |
FAM214A
|
family with sequence similarity 214, member A |
| chr12_-_68696652 | 0.07 |
ENST00000539972.1
|
MDM1
|
Mdm1 nuclear protein homolog (mouse) |
| chr6_+_168418553 | 0.07 |
ENST00000354419.2
ENST00000351261.3 |
KIF25
|
kinesin family member 25 |
| chr2_+_162165038 | 0.07 |
ENST00000437630.1
|
PSMD14
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 14 |
| chr9_+_4662282 | 0.06 |
ENST00000381883.2
|
PPAPDC2
|
phosphatidic acid phosphatase type 2 domain containing 2 |
| chr14_-_57197224 | 0.06 |
ENST00000554597.1
ENST00000556696.1 |
RP11-1085N6.3
|
Uncharacterized protein |
| chr20_+_18125727 | 0.06 |
ENST00000489634.2
|
CSRP2BP
|
CSRP2 binding protein |
| chr3_-_134092561 | 0.06 |
ENST00000510560.1
ENST00000504234.1 ENST00000515172.1 |
AMOTL2
|
angiomotin like 2 |
| chr4_-_147442817 | 0.06 |
ENST00000507030.1
|
SLC10A7
|
solute carrier family 10, member 7 |
| chrX_+_120181457 | 0.06 |
ENST00000328078.1
|
GLUD2
|
glutamate dehydrogenase 2 |
| chr3_+_112930387 | 0.06 |
ENST00000485230.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr4_+_78804393 | 0.06 |
ENST00000502384.1
|
MRPL1
|
mitochondrial ribosomal protein L1 |
| chr7_+_7811992 | 0.06 |
ENST00000406829.1
|
RPA3-AS1
|
RPA3 antisense RNA 1 |
| chr11_-_94226964 | 0.06 |
ENST00000538923.1
ENST00000540013.1 ENST00000407439.3 ENST00000393241.4 |
MRE11A
|
MRE11 meiotic recombination 11 homolog A (S. cerevisiae) |
| chrX_+_106045891 | 0.06 |
ENST00000357242.5
ENST00000310452.2 ENST00000481617.2 ENST00000276175.3 |
TBC1D8B
|
TBC1 domain family, member 8B (with GRAM domain) |
| chr1_+_95616933 | 0.06 |
ENST00000604203.1
|
RP11-57H12.6
|
TMEM56-RWDD3 readthrough |
| chr12_+_100041527 | 0.06 |
ENST00000324341.1
|
FAM71C
|
family with sequence similarity 71, member C |
| chr2_+_135809835 | 0.06 |
ENST00000264158.8
ENST00000539493.1 ENST00000442034.1 ENST00000425393.1 |
RAB3GAP1
|
RAB3 GTPase activating protein subunit 1 (catalytic) |
| chr17_+_79213039 | 0.06 |
ENST00000431388.2
|
C17orf89
|
chromosome 17 open reading frame 89 |
| chr6_-_46889694 | 0.06 |
ENST00000283296.7
ENST00000362015.4 ENST00000456426.2 |
GPR116
|
G protein-coupled receptor 116 |
| chr4_+_146403912 | 0.06 |
ENST00000507367.1
ENST00000394092.2 ENST00000515385.1 |
SMAD1
|
SMAD family member 1 |
| chr9_-_136933134 | 0.06 |
ENST00000303407.7
|
BRD3
|
bromodomain containing 3 |
| chr7_+_37723420 | 0.06 |
ENST00000476620.1
|
EPDR1
|
ependymin related 1 |
| chr7_-_37026108 | 0.06 |
ENST00000396045.3
|
ELMO1
|
engulfment and cell motility 1 |
| chr16_+_69958887 | 0.06 |
ENST00000568684.1
|
WWP2
|
WW domain containing E3 ubiquitin protein ligase 2 |
| chr9_+_108006880 | 0.06 |
ENST00000374723.1
ENST00000374720.3 ENST00000374724.1 |
SLC44A1
|
solute carrier family 44 (choline transporter), member 1 |
| chr17_-_67057047 | 0.06 |
ENST00000495634.1
ENST00000453985.2 ENST00000585714.1 |
ABCA9
|
ATP-binding cassette, sub-family A (ABC1), member 9 |
| chr5_-_64920115 | 0.06 |
ENST00000381018.3
ENST00000274327.7 |
TRIM23
|
tripartite motif containing 23 |
| chr12_-_100486668 | 0.06 |
ENST00000550544.1
ENST00000551980.1 ENST00000548045.1 ENST00000545232.2 ENST00000551973.1 |
UHRF1BP1L
|
UHRF1 binding protein 1-like |
| chr1_+_203765437 | 0.06 |
ENST00000550078.1
|
ZBED6
|
zinc finger, BED-type containing 6 |
| chr18_-_21852143 | 0.05 |
ENST00000399443.3
|
OSBPL1A
|
oxysterol binding protein-like 1A |
| chr2_-_225434538 | 0.05 |
ENST00000409096.1
|
CUL3
|
cullin 3 |
| chr11_-_77531858 | 0.05 |
ENST00000360355.2
|
RSF1
|
remodeling and spacing factor 1 |
| chr16_+_28857957 | 0.05 |
ENST00000567536.1
|
SH2B1
|
SH2B adaptor protein 1 |
| chr2_+_152266392 | 0.05 |
ENST00000444746.2
ENST00000453091.2 ENST00000428287.2 ENST00000433166.2 ENST00000420714.3 ENST00000243326.5 ENST00000414861.2 |
RIF1
|
RAP1 interacting factor homolog (yeast) |
| chr8_-_141931287 | 0.05 |
ENST00000517887.1
|
PTK2
|
protein tyrosine kinase 2 |
| chr6_+_134758827 | 0.05 |
ENST00000431422.1
|
LINC01010
|
long intergenic non-protein coding RNA 1010 |
| chr11_+_34999328 | 0.05 |
ENST00000526309.1
|
PDHX
|
pyruvate dehydrogenase complex, component X |
| chr8_+_95558771 | 0.05 |
ENST00000391679.1
|
AC023632.1
|
HCG2009141; PRO2397; Uncharacterized protein |
| chr1_+_145524891 | 0.05 |
ENST00000369304.3
|
ITGA10
|
integrin, alpha 10 |
| chr6_+_134273300 | 0.05 |
ENST00000416965.1
|
TBPL1
|
TBP-like 1 |
| chr2_+_170683979 | 0.05 |
ENST00000418381.1
|
UBR3
|
ubiquitin protein ligase E3 component n-recognin 3 (putative) |
| chr11_+_844067 | 0.05 |
ENST00000397406.1
ENST00000409543.2 ENST00000525201.1 |
TSPAN4
|
tetraspanin 4 |
| chr14_+_102606181 | 0.05 |
ENST00000335263.5
ENST00000322340.5 ENST00000424963.2 ENST00000342702.3 ENST00000556807.1 ENST00000499851.2 ENST00000558567.1 ENST00000299135.6 ENST00000454394.2 ENST00000556511.2 |
WDR20
|
WD repeat domain 20 |
| chr11_+_77532155 | 0.05 |
ENST00000532481.1
ENST00000526415.1 ENST00000393427.2 ENST00000527134.1 ENST00000304716.8 |
AAMDC
|
adipogenesis associated, Mth938 domain containing |
| chr10_-_25010795 | 0.05 |
ENST00000416305.1
ENST00000376410.2 |
ARHGAP21
|
Rho GTPase activating protein 21 |
| chr3_-_58522880 | 0.05 |
ENST00000474098.1
|
ACOX2
|
acyl-CoA oxidase 2, branched chain |
| chr17_-_8263538 | 0.05 |
ENST00000535173.1
|
AC135178.1
|
HCG1985372; Uncharacterized protein; cDNA FLJ37541 fis, clone BRCAN2026340 |
| chr5_+_60933634 | 0.05 |
ENST00000505642.1
|
C5orf64
|
chromosome 5 open reading frame 64 |
| chr10_+_123923205 | 0.05 |
ENST00000369004.3
ENST00000260733.3 |
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr6_-_42016385 | 0.05 |
ENST00000502771.1
ENST00000508143.1 ENST00000514588.1 ENST00000510503.1 ENST00000415497.2 ENST00000372988.4 |
CCND3
|
cyclin D3 |
| chr11_-_94227029 | 0.05 |
ENST00000323977.3
ENST00000536754.1 ENST00000323929.3 |
MRE11A
|
MRE11 meiotic recombination 11 homolog A (S. cerevisiae) |
| chr12_+_121416340 | 0.05 |
ENST00000257555.6
ENST00000400024.2 |
HNF1A
|
HNF1 homeobox A |
| chr14_+_96968707 | 0.05 |
ENST00000216277.8
ENST00000557320.1 ENST00000557471.1 |
PAPOLA
|
poly(A) polymerase alpha |
| chr3_-_28390298 | 0.05 |
ENST00000457172.1
|
AZI2
|
5-azacytidine induced 2 |
| chr10_+_86184676 | 0.05 |
ENST00000543283.1
|
CCSER2
|
coiled-coil serine-rich protein 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXO4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0035623 | renal glucose absorption(GO:0035623) |
| 0.1 | 0.3 | GO:0034402 | recruitment of 3'-end processing factors to RNA polymerase II holoenzyme complex(GO:0034402) |
| 0.1 | 0.3 | GO:0031455 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.1 | 0.4 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.1 | 0.2 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.1 | 0.4 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 0.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 0.2 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.0 | 0.4 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 0.1 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.0 | 0.2 | GO:1903615 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.0 | 0.2 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.2 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.4 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.2 | GO:2001151 | regulation of renal water transport(GO:2001151) positive regulation of renal water transport(GO:2001153) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.2 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.0 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.0 | 0.1 | GO:0046603 | negative regulation of mitotic centrosome separation(GO:0046603) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.1 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.1 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) |
| 0.0 | 0.1 | GO:1903371 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.0 | 0.1 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.1 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.0 | GO:0098976 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.0 | 0.1 | GO:1902962 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.1 | GO:0042426 | choline catabolic process(GO:0042426) |
| 0.0 | 0.0 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.1 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.0 | 0.2 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.2 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.1 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.0 | GO:0010983 | positive regulation of high-density lipoprotein particle clearance(GO:0010983) |
| 0.0 | 0.1 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.0 | 0.1 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.0 | 0.1 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.1 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.0 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 0.4 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 0.2 | GO:0031213 | RSF complex(GO:0031213) |
| 0.0 | 0.1 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.0 | 0.1 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.1 | GO:0005712 | chiasma(GO:0005712) |
| 0.0 | 0.1 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 0.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.4 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:0102007 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.1 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.2 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.1 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 0.1 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.1 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 0.0 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.1 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.2 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.2 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.3 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |