Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
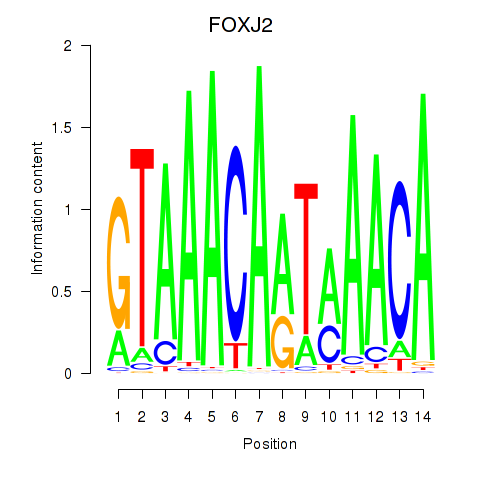
Results for FOXJ2
Z-value: 0.52
Transcription factors associated with FOXJ2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXJ2
|
ENSG00000065970.4 | forkhead box J2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXJ2 | hg19_v2_chr12_+_8185288_8185339 | 0.56 | 4.4e-01 | Click! |
Activity profile of FOXJ2 motif
Sorted Z-values of FOXJ2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_148939598 | 1.13 |
ENST00000455472.3
|
CP
|
ceruloplasmin (ferroxidase) |
| chr2_+_179318295 | 0.37 |
ENST00000442710.1
|
DFNB59
|
deafness, autosomal recessive 59 |
| chr3_-_122102065 | 0.37 |
ENST00000479899.1
ENST00000291458.5 ENST00000497726.1 |
CCDC58
|
coiled-coil domain containing 58 |
| chr4_+_57774042 | 0.34 |
ENST00000309042.7
|
REST
|
RE1-silencing transcription factor |
| chr8_+_86999516 | 0.31 |
ENST00000521564.1
|
ATP6V0D2
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2 |
| chr17_-_64225508 | 0.30 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr18_-_3013307 | 0.30 |
ENST00000584294.1
|
LPIN2
|
lipin 2 |
| chr3_-_148939835 | 0.28 |
ENST00000264613.6
|
CP
|
ceruloplasmin (ferroxidase) |
| chr8_-_95274536 | 0.28 |
ENST00000297596.2
ENST00000396194.2 |
GEM
|
GTP binding protein overexpressed in skeletal muscle |
| chr6_-_27858570 | 0.25 |
ENST00000359303.2
|
HIST1H3J
|
histone cluster 1, H3j |
| chr3_+_178866199 | 0.25 |
ENST00000263967.3
|
PIK3CA
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha |
| chr1_-_197115818 | 0.25 |
ENST00000367409.4
ENST00000294732.7 |
ASPM
|
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
| chr10_+_28822236 | 0.22 |
ENST00000347934.4
ENST00000354911.4 |
WAC
|
WW domain containing adaptor with coiled-coil |
| chr6_-_52628271 | 0.22 |
ENST00000493422.1
|
GSTA2
|
glutathione S-transferase alpha 2 |
| chr6_-_10747802 | 0.21 |
ENST00000606522.1
ENST00000606652.1 |
RP11-421M1.8
|
RP11-421M1.8 |
| chr17_+_4692230 | 0.21 |
ENST00000331264.7
|
GLTPD2
|
glycolipid transfer protein domain containing 2 |
| chr12_+_79258444 | 0.21 |
ENST00000261205.4
|
SYT1
|
synaptotagmin I |
| chr8_+_101170134 | 0.20 |
ENST00000520643.1
|
SPAG1
|
sperm associated antigen 1 |
| chr14_+_90863364 | 0.19 |
ENST00000447653.3
ENST00000553542.1 |
CALM1
|
calmodulin 1 (phosphorylase kinase, delta) |
| chr10_+_28822417 | 0.19 |
ENST00000428935.1
ENST00000420266.1 |
WAC
|
WW domain containing adaptor with coiled-coil |
| chr12_+_1099675 | 0.19 |
ENST00000545318.2
|
ERC1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr14_-_54418598 | 0.19 |
ENST00000609748.1
ENST00000558961.1 |
BMP4
|
bone morphogenetic protein 4 |
| chr2_-_55237484 | 0.19 |
ENST00000394609.2
|
RTN4
|
reticulon 4 |
| chr12_+_21679220 | 0.18 |
ENST00000256969.2
|
C12orf39
|
chromosome 12 open reading frame 39 |
| chr9_+_5890802 | 0.17 |
ENST00000381477.3
ENST00000381476.1 ENST00000381471.1 |
MLANA
|
melan-A |
| chr2_+_120770686 | 0.16 |
ENST00000331393.4
ENST00000443124.1 |
EPB41L5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr1_-_217804377 | 0.16 |
ENST00000366935.3
ENST00000366934.3 |
GPATCH2
|
G patch domain containing 2 |
| chr20_-_34638841 | 0.16 |
ENST00000565493.1
|
LINC00657
|
long intergenic non-protein coding RNA 657 |
| chr8_+_101170257 | 0.16 |
ENST00000251809.3
|
SPAG1
|
sperm associated antigen 1 |
| chr6_+_26124373 | 0.16 |
ENST00000377791.2
ENST00000602637.1 |
HIST1H2AC
|
histone cluster 1, H2ac |
| chr5_-_10308125 | 0.16 |
ENST00000296658.3
|
CMBL
|
carboxymethylenebutenolidase homolog (Pseudomonas) |
| chr17_-_79838860 | 0.16 |
ENST00000582866.1
|
RP11-498C9.15
|
RP11-498C9.15 |
| chrX_+_108779004 | 0.16 |
ENST00000218004.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chr13_-_37573432 | 0.16 |
ENST00000413537.2
ENST00000443765.1 ENST00000239891.3 |
ALG5
|
ALG5, dolichyl-phosphate beta-glucosyltransferase |
| chr8_+_104383728 | 0.15 |
ENST00000330295.5
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr2_+_120770581 | 0.15 |
ENST00000263713.5
|
EPB41L5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr3_+_132036207 | 0.15 |
ENST00000336375.5
ENST00000495911.1 |
ACPP
|
acid phosphatase, prostate |
| chr15_+_66797627 | 0.15 |
ENST00000565627.1
ENST00000564179.1 |
ZWILCH
|
zwilch kinetochore protein |
| chr3_+_127317945 | 0.15 |
ENST00000472731.1
|
MCM2
|
minichromosome maintenance complex component 2 |
| chr5_-_138780159 | 0.15 |
ENST00000512473.1
ENST00000515581.1 ENST00000515277.1 |
DNAJC18
|
DnaJ (Hsp40) homolog, subfamily C, member 18 |
| chr13_-_96329048 | 0.15 |
ENST00000606011.1
ENST00000499499.2 |
DNAJC3-AS1
|
DNAJC3 antisense RNA 1 (head to head) |
| chrX_+_102024075 | 0.14 |
ENST00000431616.1
ENST00000440496.1 ENST00000420471.1 ENST00000435966.1 |
LINC00630
|
long intergenic non-protein coding RNA 630 |
| chr15_+_66797455 | 0.14 |
ENST00000446801.2
|
ZWILCH
|
zwilch kinetochore protein |
| chr3_+_132036243 | 0.13 |
ENST00000475741.1
ENST00000351273.7 |
ACPP
|
acid phosphatase, prostate |
| chr5_-_127418573 | 0.13 |
ENST00000508353.1
ENST00000508878.1 ENST00000501652.1 ENST00000514409.1 |
CTC-228N24.3
|
CTC-228N24.3 |
| chr7_+_72742178 | 0.13 |
ENST00000442793.1
ENST00000413573.2 ENST00000252037.4 |
FKBP6
|
FK506 binding protein 6, 36kDa |
| chr6_-_56507586 | 0.13 |
ENST00000439203.1
ENST00000518935.1 ENST00000446842.2 ENST00000370765.6 ENST00000244364.6 |
DST
|
dystonin |
| chr4_-_38806404 | 0.13 |
ENST00000308979.2
ENST00000505940.1 ENST00000515861.1 |
TLR1
|
toll-like receptor 1 |
| chr11_-_5537920 | 0.13 |
ENST00000380184.1
|
UBQLNL
|
ubiquilin-like |
| chr6_+_31082603 | 0.13 |
ENST00000259881.9
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1 |
| chr3_-_57233966 | 0.13 |
ENST00000473921.1
ENST00000295934.3 |
HESX1
|
HESX homeobox 1 |
| chr10_-_25010795 | 0.12 |
ENST00000416305.1
ENST00000376410.2 |
ARHGAP21
|
Rho GTPase activating protein 21 |
| chr7_+_23637763 | 0.12 |
ENST00000410069.1
|
CCDC126
|
coiled-coil domain containing 126 |
| chr4_-_99851766 | 0.12 |
ENST00000450253.2
|
EIF4E
|
eukaryotic translation initiation factor 4E |
| chr8_-_80993010 | 0.12 |
ENST00000537855.1
ENST00000520527.1 ENST00000517427.1 ENST00000448733.2 ENST00000379097.3 |
TPD52
|
tumor protein D52 |
| chr1_+_145883868 | 0.12 |
ENST00000447947.2
|
GPR89C
|
G protein-coupled receptor 89C |
| chrX_+_55744228 | 0.12 |
ENST00000262850.7
|
RRAGB
|
Ras-related GTP binding B |
| chr7_+_95171457 | 0.11 |
ENST00000601424.1
|
AC002451.1
|
Protein LOC100996577 |
| chr6_+_76599809 | 0.11 |
ENST00000430435.1
|
MYO6
|
myosin VI |
| chr7_+_108210012 | 0.11 |
ENST00000249356.3
|
DNAJB9
|
DnaJ (Hsp40) homolog, subfamily B, member 9 |
| chr18_-_21852143 | 0.11 |
ENST00000399443.3
|
OSBPL1A
|
oxysterol binding protein-like 1A |
| chr1_+_223889285 | 0.11 |
ENST00000433674.2
|
CAPN2
|
calpain 2, (m/II) large subunit |
| chr13_+_28813645 | 0.10 |
ENST00000282391.5
|
PAN3
|
PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr10_+_28822636 | 0.10 |
ENST00000442148.1
ENST00000448193.1 |
WAC
|
WW domain containing adaptor with coiled-coil |
| chr19_+_2819854 | 0.10 |
ENST00000317243.5
|
ZNF554
|
zinc finger protein 554 |
| chr9_+_110046334 | 0.10 |
ENST00000416373.2
|
RAD23B
|
RAD23 homolog B (S. cerevisiae) |
| chr17_-_40215075 | 0.10 |
ENST00000436535.3
|
ZNF385C
|
zinc finger protein 385C |
| chrX_-_30877837 | 0.10 |
ENST00000378930.3
|
TAB3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chr21_-_35899113 | 0.10 |
ENST00000492600.1
ENST00000481448.1 ENST00000381132.2 |
RCAN1
|
regulator of calcineurin 1 |
| chr11_+_6226782 | 0.09 |
ENST00000316375.2
|
C11orf42
|
chromosome 11 open reading frame 42 |
| chr9_+_4662282 | 0.09 |
ENST00000381883.2
|
PPAPDC2
|
phosphatidic acid phosphatase type 2 domain containing 2 |
| chr1_+_93544791 | 0.09 |
ENST00000545708.1
ENST00000540243.1 ENST00000370298.4 |
MTF2
|
metal response element binding transcription factor 2 |
| chr19_-_56047661 | 0.09 |
ENST00000344158.3
|
SBK2
|
SH3 domain binding kinase family, member 2 |
| chr16_-_90038866 | 0.09 |
ENST00000314994.3
|
CENPBD1
|
CENPB DNA-binding domains containing 1 |
| chr11_+_35201826 | 0.09 |
ENST00000531873.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr17_+_17685422 | 0.08 |
ENST00000395774.1
|
RAI1
|
retinoic acid induced 1 |
| chr5_-_34919094 | 0.08 |
ENST00000341754.4
|
RAD1
|
RAD1 homolog (S. pombe) |
| chrX_-_107682702 | 0.08 |
ENST00000372216.4
|
COL4A6
|
collagen, type IV, alpha 6 |
| chr4_-_164534657 | 0.08 |
ENST00000339875.5
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr15_+_42696992 | 0.08 |
ENST00000561817.1
|
CAPN3
|
calpain 3, (p94) |
| chr7_+_73242069 | 0.08 |
ENST00000435050.1
|
CLDN4
|
claudin 4 |
| chr2_+_120770645 | 0.08 |
ENST00000443902.2
|
EPB41L5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr9_+_33617760 | 0.08 |
ENST00000379435.3
|
TRBV20OR9-2
|
T cell receptor beta variable 20/OR9-2 (non-functional) |
| chr4_-_140005443 | 0.08 |
ENST00000510408.1
ENST00000420916.2 ENST00000358635.3 |
ELF2
|
E74-like factor 2 (ets domain transcription factor) |
| chr17_-_56494882 | 0.08 |
ENST00000584437.1
|
RNF43
|
ring finger protein 43 |
| chr4_-_113627966 | 0.08 |
ENST00000505632.1
|
RP11-148B6.2
|
RP11-148B6.2 |
| chr3_-_156878540 | 0.08 |
ENST00000461804.1
|
CCNL1
|
cyclin L1 |
| chr2_+_162016916 | 0.07 |
ENST00000405852.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr4_+_78804393 | 0.07 |
ENST00000502384.1
|
MRPL1
|
mitochondrial ribosomal protein L1 |
| chr6_+_56911336 | 0.07 |
ENST00000370733.4
|
KIAA1586
|
KIAA1586 |
| chrX_+_56644819 | 0.07 |
ENST00000446028.1
|
RP11-431N15.2
|
RP11-431N15.2 |
| chr2_+_162016827 | 0.07 |
ENST00000429217.1
ENST00000406287.1 ENST00000402568.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr10_+_95848824 | 0.07 |
ENST00000371385.3
ENST00000371375.1 |
PLCE1
|
phospholipase C, epsilon 1 |
| chr9_-_71869642 | 0.07 |
ENST00000600472.1
|
AL358113.1
|
Tight junction protein 2 (Zona occludens 2) isoform 1 variant; Uncharacterized protein |
| chr15_-_56757329 | 0.07 |
ENST00000260453.3
|
MNS1
|
meiosis-specific nuclear structural 1 |
| chr19_+_57751973 | 0.06 |
ENST00000535550.1
|
ZNF805
|
zinc finger protein 805 |
| chr9_+_5510558 | 0.06 |
ENST00000397747.3
|
PDCD1LG2
|
programmed cell death 1 ligand 2 |
| chrX_+_55744166 | 0.06 |
ENST00000374941.4
ENST00000414239.1 |
RRAGB
|
Ras-related GTP binding B |
| chr9_-_21142144 | 0.06 |
ENST00000380229.2
|
IFNW1
|
interferon, omega 1 |
| chr19_+_57752022 | 0.06 |
ENST00000354309.4
|
ZNF805
|
zinc finger protein 805 |
| chrX_+_108780347 | 0.06 |
ENST00000372103.1
|
NXT2
|
nuclear transport factor 2-like export factor 2 |
| chr7_-_33140498 | 0.06 |
ENST00000448915.1
|
RP9
|
retinitis pigmentosa 9 (autosomal dominant) |
| chr10_+_60936347 | 0.06 |
ENST00000373880.4
|
PHYHIPL
|
phytanoyl-CoA 2-hydroxylase interacting protein-like |
| chr16_+_2106134 | 0.06 |
ENST00000467949.1
|
TSC2
|
tuberous sclerosis 2 |
| chr6_+_33422343 | 0.06 |
ENST00000395064.2
|
ZBTB9
|
zinc finger and BTB domain containing 9 |
| chr5_+_179135246 | 0.06 |
ENST00000508787.1
|
CANX
|
calnexin |
| chr12_+_56495115 | 0.06 |
ENST00000548861.1
|
RP11-603J24.9
|
Uncharacterized protein |
| chr1_+_95616933 | 0.05 |
ENST00000604203.1
|
RP11-57H12.6
|
TMEM56-RWDD3 readthrough |
| chr15_-_34628951 | 0.05 |
ENST00000397707.2
ENST00000560611.1 |
SLC12A6
|
solute carrier family 12 (potassium/chloride transporter), member 6 |
| chr21_+_40752378 | 0.05 |
ENST00000398753.1
ENST00000442773.1 |
WRB
|
tryptophan rich basic protein |
| chr7_-_80548493 | 0.05 |
ENST00000536800.1
|
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr3_+_160822462 | 0.05 |
ENST00000468606.1
ENST00000460503.1 |
NMD3
|
NMD3 ribosome export adaptor |
| chr2_+_231921574 | 0.05 |
ENST00000308696.6
ENST00000373635.4 ENST00000440838.1 ENST00000409643.1 |
PSMD1
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 1 |
| chr17_-_56494908 | 0.05 |
ENST00000577716.1
|
RNF43
|
ring finger protein 43 |
| chr1_+_81106951 | 0.05 |
ENST00000443565.1
|
RP5-887A10.1
|
RP5-887A10.1 |
| chr1_-_27701307 | 0.05 |
ENST00000270879.4
ENST00000354982.2 |
FCN3
|
ficolin (collagen/fibrinogen domain containing) 3 |
| chr15_+_42697065 | 0.05 |
ENST00000565559.1
|
CAPN3
|
calpain 3, (p94) |
| chr4_+_128554081 | 0.05 |
ENST00000335251.6
ENST00000296461.5 |
INTU
|
inturned planar cell polarity protein |
| chr19_+_57752200 | 0.05 |
ENST00000414468.2
|
ZNF805
|
zinc finger protein 805 |
| chr16_+_53469525 | 0.04 |
ENST00000544405.2
|
RBL2
|
retinoblastoma-like 2 (p130) |
| chr17_+_39346139 | 0.04 |
ENST00000398470.1
ENST00000318329.5 |
KRTAP9-1
|
keratin associated protein 9-1 |
| chr1_-_220263096 | 0.04 |
ENST00000463953.1
ENST00000354807.3 ENST00000414869.2 ENST00000498237.2 ENST00000498791.2 ENST00000544404.1 ENST00000480959.2 ENST00000322067.7 |
BPNT1
|
3'(2'), 5'-bisphosphate nucleotidase 1 |
| chr10_-_105992059 | 0.04 |
ENST00000369720.1
ENST00000369719.1 ENST00000357060.3 ENST00000428666.1 ENST00000278064.2 |
WDR96
|
WD repeat domain 96 |
| chr6_-_121655552 | 0.04 |
ENST00000275159.6
|
TBC1D32
|
TBC1 domain family, member 32 |
| chr21_-_37451680 | 0.04 |
ENST00000399201.1
|
SETD4
|
SET domain containing 4 |
| chr12_-_96429423 | 0.04 |
ENST00000228740.2
|
LTA4H
|
leukotriene A4 hydrolase |
| chr12_-_92536433 | 0.04 |
ENST00000551563.2
ENST00000546975.1 ENST00000549802.1 |
C12orf79
|
chromosome 12 open reading frame 79 |
| chr11_+_827248 | 0.04 |
ENST00000527089.1
ENST00000530183.1 |
EFCAB4A
|
EF-hand calcium binding domain 4A |
| chr19_+_47538560 | 0.04 |
ENST00000439365.2
ENST00000594670.1 |
NPAS1
|
neuronal PAS domain protein 1 |
| chr7_-_80548667 | 0.04 |
ENST00000265361.3
|
SEMA3C
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr20_+_55926625 | 0.04 |
ENST00000452119.1
|
RAE1
|
ribonucleic acid export 1 |
| chr6_+_10748019 | 0.04 |
ENST00000543878.1
ENST00000461342.1 ENST00000475942.1 ENST00000379530.3 ENST00000473276.1 ENST00000481240.1 ENST00000467317.1 |
SYCP2L
TMEM14B
|
synaptonemal complex protein 2-like transmembrane protein 14B |
| chr1_-_38019878 | 0.04 |
ENST00000296215.6
|
SNIP1
|
Smad nuclear interacting protein 1 |
| chr6_+_11094266 | 0.04 |
ENST00000416247.2
|
SMIM13
|
small integral membrane protein 13 |
| chr4_-_122148620 | 0.04 |
ENST00000509841.1
|
TNIP3
|
TNFAIP3 interacting protein 3 |
| chr20_+_23420322 | 0.04 |
ENST00000347397.1
|
CSTL1
|
cystatin-like 1 |
| chr16_+_86609939 | 0.04 |
ENST00000593625.1
|
FOXL1
|
forkhead box L1 |
| chrX_+_107683096 | 0.03 |
ENST00000328300.6
ENST00000361603.2 |
COL4A5
|
collagen, type IV, alpha 5 |
| chr1_+_221054584 | 0.03 |
ENST00000549319.1
|
HLX
|
H2.0-like homeobox |
| chr2_-_65593784 | 0.03 |
ENST00000443619.2
|
SPRED2
|
sprouty-related, EVH1 domain containing 2 |
| chr11_+_118398178 | 0.03 |
ENST00000302783.4
ENST00000539546.1 |
TTC36
|
tetratricopeptide repeat domain 36 |
| chrX_+_22056165 | 0.03 |
ENST00000535894.1
|
PHEX
|
phosphate regulating endopeptidase homolog, X-linked |
| chr7_+_77469439 | 0.03 |
ENST00000450574.1
ENST00000416283.2 ENST00000248550.7 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr11_-_57089774 | 0.03 |
ENST00000527207.1
|
TNKS1BP1
|
tankyrase 1 binding protein 1, 182kDa |
| chr12_-_7596735 | 0.03 |
ENST00000416109.2
ENST00000396630.1 ENST00000313599.3 |
CD163L1
|
CD163 molecule-like 1 |
| chr17_-_2117600 | 0.03 |
ENST00000572369.1
|
SMG6
|
SMG6 nonsense mediated mRNA decay factor |
| chr3_+_128444965 | 0.03 |
ENST00000265062.3
|
RAB7A
|
RAB7A, member RAS oncogene family |
| chr3_-_64009658 | 0.03 |
ENST00000394431.2
|
PSMD6
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 |
| chr10_-_36813162 | 0.03 |
ENST00000440465.1
|
NAMPTL
|
nicotinamide phosphoribosyltransferase-like |
| chr19_+_2819943 | 0.03 |
ENST00000591265.1
|
ZNF554
|
zinc finger protein 554 |
| chr7_-_32529973 | 0.03 |
ENST00000410044.1
ENST00000409987.1 ENST00000409782.1 ENST00000450169.2 |
LSM5
|
LSM5 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr9_-_99417562 | 0.03 |
ENST00000375234.3
ENST00000446045.1 |
AAED1
|
AhpC/TSA antioxidant enzyme domain containing 1 |
| chr2_+_162016804 | 0.03 |
ENST00000392749.2
ENST00000440506.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr11_-_57417405 | 0.03 |
ENST00000524669.1
ENST00000300022.3 |
YPEL4
|
yippee-like 4 (Drosophila) |
| chr9_+_5510492 | 0.02 |
ENST00000397745.2
|
PDCD1LG2
|
programmed cell death 1 ligand 2 |
| chr19_+_49866331 | 0.02 |
ENST00000597873.1
|
DKKL1
|
dickkopf-like 1 |
| chr14_+_90863327 | 0.02 |
ENST00000356978.4
|
CALM1
|
calmodulin 1 (phosphorylase kinase, delta) |
| chr9_-_86571628 | 0.02 |
ENST00000376344.3
|
C9orf64
|
chromosome 9 open reading frame 64 |
| chr12_+_27863706 | 0.02 |
ENST00000081029.3
ENST00000538315.1 ENST00000542791.1 |
MRPS35
|
mitochondrial ribosomal protein S35 |
| chr8_+_128426535 | 0.02 |
ENST00000465342.2
|
POU5F1B
|
POU class 5 homeobox 1B |
| chr18_-_73967160 | 0.02 |
ENST00000579714.1
|
RP11-94B19.7
|
RP11-94B19.7 |
| chr2_+_61372226 | 0.02 |
ENST00000426997.1
|
C2orf74
|
chromosome 2 open reading frame 74 |
| chr8_+_58890917 | 0.01 |
ENST00000522992.1
|
RP11-1112C15.1
|
RP11-1112C15.1 |
| chr1_+_91966384 | 0.01 |
ENST00000430031.2
ENST00000234626.6 |
CDC7
|
cell division cycle 7 |
| chr4_+_57845024 | 0.01 |
ENST00000431623.2
ENST00000441246.2 |
POLR2B
|
polymerase (RNA) II (DNA directed) polypeptide B, 140kDa |
| chr1_+_36335351 | 0.01 |
ENST00000373206.1
|
AGO1
|
argonaute RISC catalytic component 1 |
| chr9_-_134615443 | 0.01 |
ENST00000372195.1
|
RAPGEF1
|
Rap guanine nucleotide exchange factor (GEF) 1 |
| chr12_+_27396901 | 0.01 |
ENST00000541191.1
ENST00000389032.3 |
STK38L
|
serine/threonine kinase 38 like |
| chr2_-_85641162 | 0.01 |
ENST00000447219.2
ENST00000409670.1 ENST00000409724.1 |
CAPG
|
capping protein (actin filament), gelsolin-like |
| chr1_+_239882842 | 0.01 |
ENST00000448020.1
|
CHRM3
|
cholinergic receptor, muscarinic 3 |
| chr17_-_46115122 | 0.01 |
ENST00000006101.4
|
COPZ2
|
coatomer protein complex, subunit zeta 2 |
| chr11_+_63137251 | 0.01 |
ENST00000310969.4
ENST00000279178.3 |
SLC22A9
|
solute carrier family 22 (organic anion transporter), member 9 |
| chrX_-_19533379 | 0.01 |
ENST00000338883.4
|
MAP3K15
|
mitogen-activated protein kinase kinase kinase 15 |
| chr7_-_102283238 | 0.01 |
ENST00000340457.8
|
UPK3BL
|
uroplakin 3B-like |
| chr19_+_56989485 | 0.01 |
ENST00000585445.1
ENST00000586091.1 ENST00000594783.1 ENST00000592146.1 ENST00000588158.1 ENST00000299997.4 ENST00000591797.1 |
ZNF667-AS1
|
ZNF667 antisense RNA 1 (head to head) |
| chr6_-_99842041 | 0.01 |
ENST00000254759.3
ENST00000369242.1 |
COQ3
|
coenzyme Q3 methyltransferase |
| chr11_+_61197572 | 0.01 |
ENST00000542074.1
ENST00000534878.1 ENST00000537782.1 ENST00000543265.1 |
SDHAF2
|
succinate dehydrogenase complex assembly factor 2 |
| chr15_+_42697018 | 0.01 |
ENST00000397204.4
|
CAPN3
|
calpain 3, (p94) |
| chr3_+_157154578 | 0.01 |
ENST00000295927.3
|
PTX3
|
pentraxin 3, long |
| chr8_-_54755789 | 0.01 |
ENST00000359530.2
|
ATP6V1H
|
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H |
| chr9_+_131683174 | 0.01 |
ENST00000372592.3
ENST00000428610.1 |
PHYHD1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr12_-_102874102 | 0.01 |
ENST00000392905.2
|
IGF1
|
insulin-like growth factor 1 (somatomedin C) |
| chr10_+_90346519 | 0.00 |
ENST00000371939.3
|
LIPJ
|
lipase, family member J |
| chr2_-_24346218 | 0.00 |
ENST00000436622.1
ENST00000313213.4 |
PFN4
|
profilin family, member 4 |
| chr10_-_32667660 | 0.00 |
ENST00000375110.2
|
EPC1
|
enhancer of polycomb homolog 1 (Drosophila) |
| chr1_+_160765919 | 0.00 |
ENST00000341032.4
ENST00000368041.2 ENST00000368040.1 |
LY9
|
lymphocyte antigen 9 |
| chr8_-_82608409 | 0.00 |
ENST00000518568.1
|
SLC10A5
|
solute carrier family 10, member 5 |
| chr1_-_240775447 | 0.00 |
ENST00000318160.4
|
GREM2
|
gremlin 2, DAN family BMP antagonist |
| chr2_-_206950781 | 0.00 |
ENST00000403263.1
|
INO80D
|
INO80 complex subunit D |
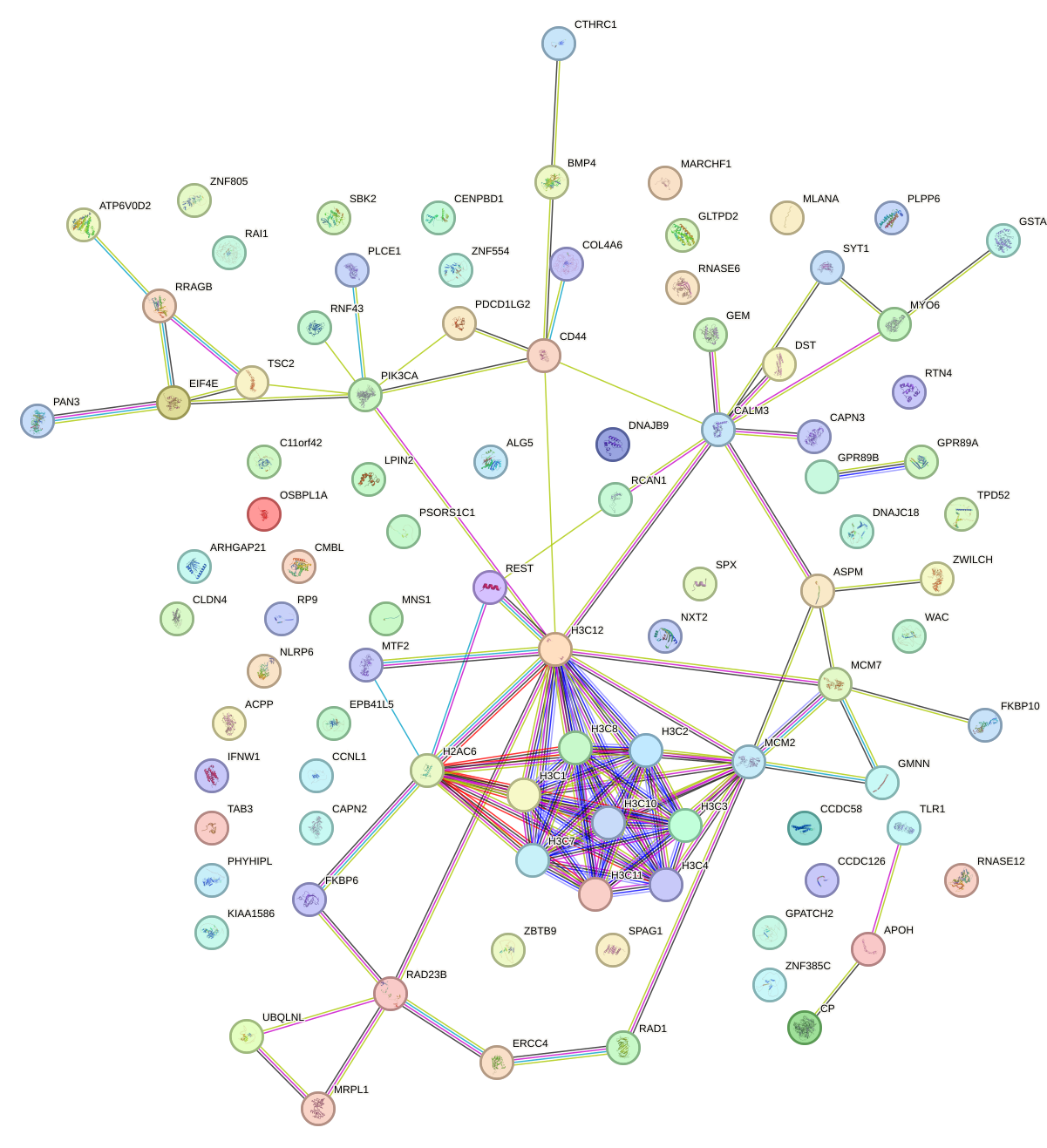
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXJ2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0071894 | histone H2B conserved C-terminal lysine ubiquitination(GO:0071894) |
| 0.1 | 0.3 | GO:2000705 | dense core granule biogenesis(GO:0061110) regulation of dense core granule biogenesis(GO:2000705) |
| 0.1 | 0.3 | GO:0060168 | positive regulation of adenosine receptor signaling pathway(GO:0060168) |
| 0.1 | 0.3 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.3 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.1 | 0.3 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.1 | 0.4 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.0 | 0.1 | GO:0042495 | detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 1.4 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.2 | GO:0061150 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.0 | 0.2 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.0 | 0.3 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.1 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.2 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.1 | GO:0051598 | meiotic recombination checkpoint(GO:0051598) |
| 0.0 | 0.1 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.0 | 0.2 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.2 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.1 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.1 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.1 | GO:0060174 | limb bud formation(GO:0060174) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.0 | 0.1 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.0 | 0.2 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.2 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.3 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.3 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.1 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.1 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.3 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.3 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.0 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.0 | 0.1 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.1 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.1 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.3 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.3 | GO:0052642 | lysophosphatidic acid phosphatase activity(GO:0052642) |
| 0.0 | 0.3 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0060001 | minus-end directed microfilament motor activity(GO:0060001) |
| 0.0 | 0.1 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.0 | 0.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.3 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.3 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.2 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.2 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.0 | GO:0008441 | 3'(2'),5'-bisphosphate nucleotidase activity(GO:0008441) |
| 0.0 | 0.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.5 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.2 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.1 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.1 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.2 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |