Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
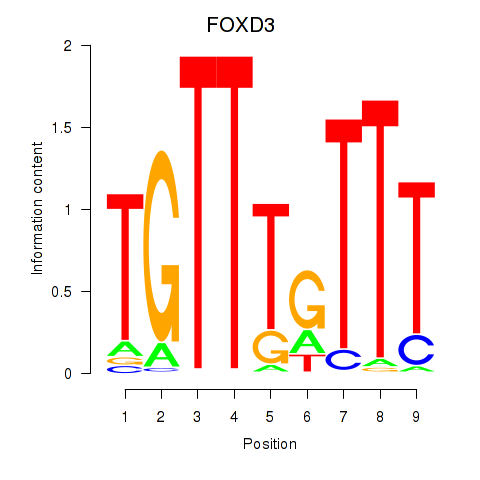
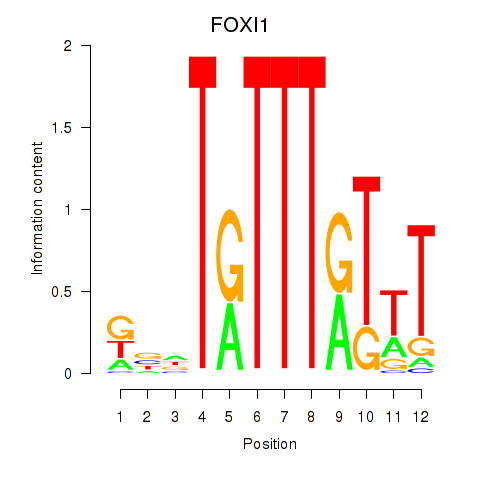
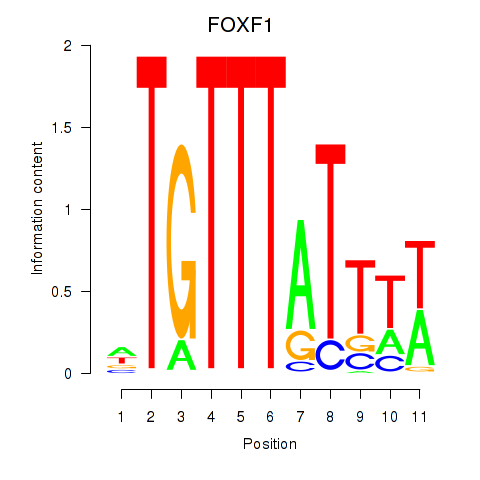
Results for FOXD3_FOXI1_FOXF1
Z-value: 1.10
Transcription factors associated with FOXD3_FOXI1_FOXF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXD3
|
ENSG00000187140.4 | forkhead box D3 |
|
FOXI1
|
ENSG00000168269.7 | forkhead box I1 |
|
FOXF1
|
ENSG00000103241.5 | forkhead box F1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXD3 | hg19_v2_chr1_+_63788730_63788730 | 0.87 | 1.3e-01 | Click! |
Activity profile of FOXD3_FOXI1_FOXF1 motif
Sorted Z-values of FOXD3_FOXI1_FOXF1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_40841410 | 1.03 |
ENST00000381677.3
|
CARD6
|
caspase recruitment domain family, member 6 |
| chr9_+_78505581 | 1.03 |
ENST00000376767.3
ENST00000376752.4 |
PCSK5
|
proprotein convertase subtilisin/kexin type 5 |
| chr2_+_232575168 | 0.93 |
ENST00000440384.1
|
PTMA
|
prothymosin, alpha |
| chr1_+_207277632 | 0.86 |
ENST00000421786.1
|
C4BPA
|
complement component 4 binding protein, alpha |
| chr8_+_86999516 | 0.76 |
ENST00000521564.1
|
ATP6V0D2
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2 |
| chr3_-_148939598 | 0.72 |
ENST00000455472.3
|
CP
|
ceruloplasmin (ferroxidase) |
| chr15_-_76352069 | 0.68 |
ENST00000305435.10
ENST00000563910.1 |
NRG4
|
neuregulin 4 |
| chr8_+_52730143 | 0.64 |
ENST00000415643.1
|
AC090186.1
|
Uncharacterized protein |
| chr17_+_57408994 | 0.63 |
ENST00000312655.4
|
YPEL2
|
yippee-like 2 (Drosophila) |
| chr6_-_134861089 | 0.59 |
ENST00000606039.1
|
RP11-557H15.4
|
RP11-557H15.4 |
| chr10_-_14050522 | 0.56 |
ENST00000342409.2
|
FRMD4A
|
FERM domain containing 4A |
| chr9_-_100000957 | 0.54 |
ENST00000366109.2
ENST00000607322.1 |
RP11-498P14.5
|
RP11-498P14.5 |
| chr14_+_51026844 | 0.50 |
ENST00000554886.1
|
ATL1
|
atlastin GTPase 1 |
| chr3_+_119316721 | 0.50 |
ENST00000488919.1
ENST00000495992.1 |
PLA1A
|
phospholipase A1 member A |
| chr18_-_52626622 | 0.50 |
ENST00000591504.1
|
CCDC68
|
coiled-coil domain containing 68 |
| chr3_-_193096600 | 0.46 |
ENST00000446087.1
ENST00000342358.4 |
ATP13A5
|
ATPase type 13A5 |
| chr9_-_4859260 | 0.45 |
ENST00000599351.1
|
AL158147.2
|
HCG2011465; Uncharacterized protein |
| chr2_+_155554797 | 0.44 |
ENST00000295101.2
|
KCNJ3
|
potassium inwardly-rectifying channel, subfamily J, member 3 |
| chr6_+_125524785 | 0.43 |
ENST00000392482.2
|
TPD52L1
|
tumor protein D52-like 1 |
| chr3_+_178866199 | 0.43 |
ENST00000263967.3
|
PIK3CA
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha |
| chr5_-_59064458 | 0.42 |
ENST00000502575.1
ENST00000507116.1 |
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr21_+_39628655 | 0.41 |
ENST00000398925.1
ENST00000398928.1 ENST00000328656.4 ENST00000443341.1 |
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr5_+_125706998 | 0.40 |
ENST00000506445.1
|
GRAMD3
|
GRAM domain containing 3 |
| chr13_-_86373536 | 0.40 |
ENST00000400286.2
|
SLITRK6
|
SLIT and NTRK-like family, member 6 |
| chr15_-_38519066 | 0.40 |
ENST00000561320.1
ENST00000561161.1 |
RP11-346D14.1
|
RP11-346D14.1 |
| chr4_-_153303658 | 0.39 |
ENST00000296555.5
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr1_+_239882842 | 0.39 |
ENST00000448020.1
|
CHRM3
|
cholinergic receptor, muscarinic 3 |
| chr17_-_40215075 | 0.38 |
ENST00000436535.3
|
ZNF385C
|
zinc finger protein 385C |
| chr15_+_96869165 | 0.37 |
ENST00000421109.2
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr3_+_119316689 | 0.36 |
ENST00000273371.4
|
PLA1A
|
phospholipase A1 member A |
| chr14_+_51026926 | 0.35 |
ENST00000557735.1
|
ATL1
|
atlastin GTPase 1 |
| chr7_+_106415457 | 0.35 |
ENST00000490162.2
ENST00000470135.1 |
RP5-884M6.1
|
RP5-884M6.1 |
| chr14_+_77843459 | 0.34 |
ENST00000216471.4
|
SAMD15
|
sterile alpha motif domain containing 15 |
| chr12_+_27849378 | 0.34 |
ENST00000310791.2
|
REP15
|
RAB15 effector protein |
| chr2_-_178129551 | 0.34 |
ENST00000430047.1
|
NFE2L2
|
nuclear factor, erythroid 2-like 2 |
| chr5_+_72509751 | 0.33 |
ENST00000515556.1
ENST00000513379.1 ENST00000427584.2 |
RP11-60A8.1
|
RP11-60A8.1 |
| chr4_+_76871883 | 0.33 |
ENST00000599764.1
|
AC110615.1
|
Uncharacterized protein |
| chr11_+_107650219 | 0.33 |
ENST00000398067.1
|
AP001024.1
|
Uncharacterized protein |
| chr10_-_14613968 | 0.33 |
ENST00000488576.1
ENST00000472095.1 |
FAM107B
|
family with sequence similarity 107, member B |
| chr16_+_2570431 | 0.32 |
ENST00000563556.1
|
AMDHD2
|
amidohydrolase domain containing 2 |
| chr3_-_114035026 | 0.32 |
ENST00000570269.1
|
RP11-553L6.5
|
RP11-553L6.5 |
| chr12_-_68845417 | 0.31 |
ENST00000542875.1
|
RP11-81H14.2
|
RP11-81H14.2 |
| chr4_-_103266626 | 0.31 |
ENST00000356736.4
|
SLC39A8
|
solute carrier family 39 (zinc transporter), member 8 |
| chr17_-_57229155 | 0.30 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr9_-_3469181 | 0.30 |
ENST00000366116.2
|
AL365202.1
|
Uncharacterized protein |
| chr19_-_37701386 | 0.30 |
ENST00000527838.1
ENST00000591492.1 ENST00000532828.2 |
ZNF585B
|
zinc finger protein 585B |
| chr19_-_52531600 | 0.30 |
ENST00000356322.6
ENST00000270649.6 |
ZNF614
|
zinc finger protein 614 |
| chr4_+_95174445 | 0.30 |
ENST00000509418.1
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr4_+_71587669 | 0.30 |
ENST00000381006.3
ENST00000226328.4 |
RUFY3
|
RUN and FYVE domain containing 3 |
| chr1_-_150669604 | 0.29 |
ENST00000427665.1
ENST00000540514.1 |
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr12_-_106480587 | 0.29 |
ENST00000548902.1
|
NUAK1
|
NUAK family, SNF1-like kinase, 1 |
| chrM_+_8366 | 0.29 |
ENST00000361851.1
|
MT-ATP8
|
mitochondrially encoded ATP synthase 8 |
| chr11_-_8739383 | 0.28 |
ENST00000531060.1
|
ST5
|
suppression of tumorigenicity 5 |
| chrX_-_45710920 | 0.28 |
ENST00000456532.1
|
RP5-1158E12.3
|
RP5-1158E12.3 |
| chr6_+_53794780 | 0.28 |
ENST00000505762.1
ENST00000511369.1 ENST00000431554.2 |
MLIP
RP11-411K7.1
|
muscular LMNA-interacting protein RP11-411K7.1 |
| chr12_-_96389702 | 0.27 |
ENST00000552509.1
|
HAL
|
histidine ammonia-lyase |
| chr15_-_55790515 | 0.27 |
ENST00000448430.2
ENST00000457155.2 |
DYX1C1
|
dyslexia susceptibility 1 candidate 1 |
| chr2_-_36779411 | 0.27 |
ENST00000406220.1
|
AC007401.2
|
Uncharacterized protein |
| chr14_+_38065052 | 0.27 |
ENST00000556845.1
|
TTC6
|
tetratricopeptide repeat domain 6 |
| chr13_+_44596471 | 0.27 |
ENST00000592085.1
ENST00000591799.1 |
LINC00284
|
long intergenic non-protein coding RNA 284 |
| chrX_-_80377118 | 0.26 |
ENST00000373250.3
|
HMGN5
|
high mobility group nucleosome binding domain 5 |
| chrX_-_80377162 | 0.26 |
ENST00000430960.1
ENST00000447319.1 |
HMGN5
|
high mobility group nucleosome binding domain 5 |
| chr17_+_79369249 | 0.26 |
ENST00000574717.2
|
RP11-1055B8.6
|
Uncharacterized protein |
| chr10_+_114709999 | 0.26 |
ENST00000355995.4
ENST00000545257.1 ENST00000543371.1 ENST00000536810.1 ENST00000355717.4 ENST00000538897.1 ENST00000534894.1 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chrM_+_4431 | 0.26 |
ENST00000361453.3
|
MT-ND2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr12_-_102591604 | 0.26 |
ENST00000329406.4
|
PMCH
|
pro-melanin-concentrating hormone |
| chr12_-_68696652 | 0.26 |
ENST00000539972.1
|
MDM1
|
Mdm1 nuclear protein homolog (mouse) |
| chr12_-_57030096 | 0.26 |
ENST00000549506.1
|
BAZ2A
|
bromodomain adjacent to zinc finger domain, 2A |
| chr11_-_790060 | 0.26 |
ENST00000330106.4
|
CEND1
|
cell cycle exit and neuronal differentiation 1 |
| chr5_-_111091948 | 0.25 |
ENST00000447165.2
|
NREP
|
neuronal regeneration related protein |
| chr12_-_39734783 | 0.25 |
ENST00000552961.1
|
KIF21A
|
kinesin family member 21A |
| chr11_-_111781610 | 0.25 |
ENST00000525823.1
|
CRYAB
|
crystallin, alpha B |
| chr5_+_133859996 | 0.25 |
ENST00000512386.1
|
PHF15
|
jade family PHD finger 2 |
| chr12_+_78224667 | 0.25 |
ENST00000549464.1
|
NAV3
|
neuron navigator 3 |
| chr8_+_79428539 | 0.25 |
ENST00000352966.5
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha |
| chr17_-_38978847 | 0.25 |
ENST00000269576.5
|
KRT10
|
keratin 10 |
| chr1_+_104159999 | 0.25 |
ENST00000414303.2
ENST00000423678.1 |
AMY2A
|
amylase, alpha 2A (pancreatic) |
| chr5_+_149865377 | 0.25 |
ENST00000522491.1
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr12_-_8025623 | 0.24 |
ENST00000542782.1
ENST00000396589.2 ENST00000535266.1 ENST00000542505.1 |
SLC2A14
|
solute carrier family 2 (facilitated glucose transporter), member 14 |
| chr19_-_46627914 | 0.24 |
ENST00000341415.2
|
IGFL3
|
IGF-like family member 3 |
| chr2_-_182545603 | 0.23 |
ENST00000295108.3
|
NEUROD1
|
neuronal differentiation 1 |
| chr15_-_31521567 | 0.23 |
ENST00000560812.1
ENST00000559853.1 ENST00000558109.1 |
RP11-16E12.2
|
RP11-16E12.2 |
| chr3_+_50712672 | 0.23 |
ENST00000266037.9
|
DOCK3
|
dedicator of cytokinesis 3 |
| chr1_-_150669500 | 0.23 |
ENST00000271732.3
|
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr14_+_88490894 | 0.23 |
ENST00000556033.1
ENST00000553929.1 ENST00000555996.1 ENST00000556673.1 ENST00000557339.1 ENST00000556684.1 |
RP11-300J18.3
|
long intergenic non-protein coding RNA 1146 |
| chr3_+_189349162 | 0.23 |
ENST00000264731.3
ENST00000382063.4 ENST00000418709.2 ENST00000320472.5 ENST00000392460.3 ENST00000440651.2 |
TP63
|
tumor protein p63 |
| chr1_-_63782888 | 0.23 |
ENST00000436475.2
|
LINC00466
|
long intergenic non-protein coding RNA 466 |
| chr7_+_128312346 | 0.22 |
ENST00000480462.1
ENST00000378704.3 ENST00000477515.1 |
FAM71F2
|
family with sequence similarity 71, member F2 |
| chr2_-_44550441 | 0.22 |
ENST00000420756.1
ENST00000444696.1 |
PREPL
|
prolyl endopeptidase-like |
| chrX_+_22050546 | 0.22 |
ENST00000379374.4
|
PHEX
|
phosphate regulating endopeptidase homolog, X-linked |
| chr14_+_90863364 | 0.22 |
ENST00000447653.3
ENST00000553542.1 |
CALM1
|
calmodulin 1 (phosphorylase kinase, delta) |
| chr19_-_15443318 | 0.21 |
ENST00000360016.5
|
BRD4
|
bromodomain containing 4 |
| chr12_-_3982511 | 0.21 |
ENST00000427057.2
ENST00000228820.4 |
PARP11
|
poly (ADP-ribose) polymerase family, member 11 |
| chr7_+_142985308 | 0.21 |
ENST00000310447.5
|
CASP2
|
caspase 2, apoptosis-related cysteine peptidase |
| chr1_+_229440129 | 0.21 |
ENST00000366688.3
|
SPHAR
|
S-phase response (cyclin related) |
| chr2_+_168725458 | 0.21 |
ENST00000392690.3
|
B3GALT1
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr3_+_136649311 | 0.21 |
ENST00000469404.1
ENST00000467911.1 |
NCK1
|
NCK adaptor protein 1 |
| chr5_-_41510656 | 0.21 |
ENST00000377801.3
|
PLCXD3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr15_-_33180439 | 0.21 |
ENST00000559610.1
|
FMN1
|
formin 1 |
| chr1_+_104104379 | 0.21 |
ENST00000435302.1
|
AMY2B
|
amylase, alpha 2B (pancreatic) |
| chr12_-_21487829 | 0.21 |
ENST00000445053.1
ENST00000452078.1 ENST00000458504.1 ENST00000422327.1 ENST00000421294.1 |
SLCO1A2
|
solute carrier organic anion transporter family, member 1A2 |
| chr3_+_172034218 | 0.20 |
ENST00000366261.2
|
AC092964.1
|
Uncharacterized protein |
| chr17_+_4692230 | 0.20 |
ENST00000331264.7
|
GLTPD2
|
glycolipid transfer protein domain containing 2 |
| chr6_+_77484663 | 0.20 |
ENST00000607287.1
|
RP11-354K4.2
|
RP11-354K4.2 |
| chr4_-_140544386 | 0.20 |
ENST00000561977.1
|
RP11-308D13.3
|
RP11-308D13.3 |
| chr12_+_32655110 | 0.20 |
ENST00000546442.1
ENST00000583694.1 |
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr2_-_72374948 | 0.20 |
ENST00000546307.1
ENST00000474509.1 |
CYP26B1
|
cytochrome P450, family 26, subfamily B, polypeptide 1 |
| chr2_-_102091144 | 0.20 |
ENST00000428343.1
|
RFX8
|
RFX family member 8, lacking RFX DNA binding domain |
| chr9_+_100000717 | 0.20 |
ENST00000375205.2
ENST00000357054.1 ENST00000395220.1 ENST00000375202.2 ENST00000411667.2 |
CCDC180
|
coiled-coil domain containing 180 |
| chr19_-_13900972 | 0.19 |
ENST00000397557.1
|
AC008686.1
|
Uncharacterized protein |
| chr3_+_157827841 | 0.19 |
ENST00000295930.3
ENST00000471994.1 ENST00000464171.1 ENST00000312179.6 ENST00000475278.2 |
RSRC1
|
arginine/serine-rich coiled-coil 1 |
| chr3_-_182881595 | 0.19 |
ENST00000476015.1
|
LAMP3
|
lysosomal-associated membrane protein 3 |
| chr7_+_95171457 | 0.19 |
ENST00000601424.1
|
AC002451.1
|
Protein LOC100996577 |
| chr8_-_117886563 | 0.19 |
ENST00000519837.1
ENST00000522699.1 |
RAD21
|
RAD21 homolog (S. pombe) |
| chr14_+_102276192 | 0.19 |
ENST00000557714.1
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr14_+_31028329 | 0.19 |
ENST00000206595.6
|
G2E3
|
G2/M-phase specific E3 ubiquitin protein ligase |
| chr17_-_74547256 | 0.19 |
ENST00000589145.1
|
CYGB
|
cytoglobin |
| chr10_-_90712520 | 0.19 |
ENST00000224784.6
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta |
| chrX_+_41583408 | 0.19 |
ENST00000302548.4
|
GPR82
|
G protein-coupled receptor 82 |
| chr4_-_130014729 | 0.19 |
ENST00000281142.5
ENST00000434680.1 |
SCLT1
|
sodium channel and clathrin linker 1 |
| chr19_-_11689752 | 0.19 |
ENST00000592659.1
ENST00000592828.1 ENST00000218758.5 ENST00000412435.2 |
ACP5
|
acid phosphatase 5, tartrate resistant |
| chr15_+_58430368 | 0.19 |
ENST00000558772.1
ENST00000219919.4 |
AQP9
|
aquaporin 9 |
| chr17_-_77967433 | 0.19 |
ENST00000571872.1
|
TBC1D16
|
TBC1 domain family, member 16 |
| chrX_-_154563889 | 0.18 |
ENST00000369449.2
ENST00000321926.4 |
CLIC2
|
chloride intracellular channel 2 |
| chr2_+_13677795 | 0.18 |
ENST00000434509.1
|
AC092635.1
|
AC092635.1 |
| chr4_-_176733897 | 0.18 |
ENST00000393658.2
|
GPM6A
|
glycoprotein M6A |
| chr18_+_52258390 | 0.18 |
ENST00000321600.1
|
DYNAP
|
dynactin associated protein |
| chr22_+_40440804 | 0.18 |
ENST00000441751.1
ENST00000301923.9 |
TNRC6B
|
trinucleotide repeat containing 6B |
| chr5_-_142065612 | 0.18 |
ENST00000360966.5
ENST00000411960.1 |
FGF1
|
fibroblast growth factor 1 (acidic) |
| chr9_-_99064429 | 0.18 |
ENST00000375263.3
|
HSD17B3
|
hydroxysteroid (17-beta) dehydrogenase 3 |
| chr17_+_76311791 | 0.18 |
ENST00000586321.1
|
AC061992.2
|
AC061992.2 |
| chr4_+_155484155 | 0.18 |
ENST00000509493.1
|
FGB
|
fibrinogen beta chain |
| chr4_-_164534657 | 0.18 |
ENST00000339875.5
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr11_+_17316870 | 0.18 |
ENST00000458064.2
|
NUCB2
|
nucleobindin 2 |
| chr2_-_165698662 | 0.18 |
ENST00000194871.6
ENST00000445474.2 |
COBLL1
|
cordon-bleu WH2 repeat protein-like 1 |
| chr6_+_45296391 | 0.18 |
ENST00000371436.6
ENST00000576263.1 |
RUNX2
|
runt-related transcription factor 2 |
| chr9_+_26956474 | 0.17 |
ENST00000429045.2
|
IFT74
|
intraflagellar transport 74 homolog (Chlamydomonas) |
| chr3_+_28390637 | 0.17 |
ENST00000420223.1
ENST00000383768.2 |
ZCWPW2
|
zinc finger, CW type with PWWP domain 2 |
| chr17_-_39341594 | 0.17 |
ENST00000398472.1
|
KRTAP4-1
|
keratin associated protein 4-1 |
| chr22_-_21360736 | 0.17 |
ENST00000547793.2
|
AC002472.1
|
Uncharacterized protein |
| chr11_-_8739566 | 0.17 |
ENST00000533020.1
|
ST5
|
suppression of tumorigenicity 5 |
| chr17_-_29641104 | 0.17 |
ENST00000577894.1
ENST00000330927.4 |
EVI2B
|
ecotropic viral integration site 2B |
| chr5_+_98104978 | 0.17 |
ENST00000308234.7
|
RGMB
|
repulsive guidance molecule family member b |
| chr14_-_54418598 | 0.17 |
ENST00000609748.1
ENST00000558961.1 |
BMP4
|
bone morphogenetic protein 4 |
| chr4_-_99850243 | 0.17 |
ENST00000280892.6
ENST00000511644.1 ENST00000504432.1 ENST00000505992.1 |
EIF4E
|
eukaryotic translation initiation factor 4E |
| chr14_+_67831576 | 0.16 |
ENST00000555876.1
|
EIF2S1
|
eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa |
| chr18_+_77160282 | 0.16 |
ENST00000318065.5
ENST00000545796.1 ENST00000592223.1 ENST00000329101.4 ENST00000586434.1 |
NFATC1
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 |
| chr8_-_42360015 | 0.16 |
ENST00000522707.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr12_-_99038732 | 0.16 |
ENST00000393042.3
ENST00000420861.1 ENST00000299157.4 ENST00000342502.2 |
IKBIP
|
IKBKB interacting protein |
| chr12_+_69186125 | 0.16 |
ENST00000399333.3
|
AC124890.1
|
HCG1774533, isoform CRA_a; PRO2268; Uncharacterized protein |
| chr8_-_82754193 | 0.16 |
ENST00000519817.1
ENST00000521773.1 ENST00000523757.1 |
SNX16
|
sorting nexin 16 |
| chr5_-_142782862 | 0.16 |
ENST00000415690.2
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr19_-_23456996 | 0.16 |
ENST00000594653.1
|
RP11-15H20.7
|
RP11-15H20.7 |
| chr4_-_100140331 | 0.16 |
ENST00000407820.2
ENST00000394897.1 ENST00000508558.1 ENST00000394899.2 |
ADH6
|
alcohol dehydrogenase 6 (class V) |
| chr16_-_53737722 | 0.16 |
ENST00000569716.1
ENST00000562588.1 ENST00000562230.1 ENST00000379925.3 ENST00000563746.1 ENST00000568653.3 |
RPGRIP1L
|
RPGRIP1-like |
| chr11_-_111649015 | 0.16 |
ENST00000529841.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chrX_+_24483338 | 0.16 |
ENST00000379162.4
ENST00000441463.2 |
PDK3
|
pyruvate dehydrogenase kinase, isozyme 3 |
| chr5_-_76916396 | 0.16 |
ENST00000509971.1
|
WDR41
|
WD repeat domain 41 |
| chr4_-_99578776 | 0.16 |
ENST00000515287.1
|
TSPAN5
|
tetraspanin 5 |
| chr6_+_101846664 | 0.16 |
ENST00000421544.1
ENST00000413795.1 ENST00000369138.1 ENST00000358361.3 |
GRIK2
|
glutamate receptor, ionotropic, kainate 2 |
| chr14_-_95236551 | 0.16 |
ENST00000238558.3
|
GSC
|
goosecoid homeobox |
| chr9_+_67977438 | 0.15 |
ENST00000456982.1
|
RP11-195B21.3
|
Protein LOC644249 |
| chr1_-_98510843 | 0.15 |
ENST00000413670.2
ENST00000538428.1 |
MIR137HG
|
MIR137 host gene (non-protein coding) |
| chr7_-_27219632 | 0.15 |
ENST00000470747.4
|
RP1-170O19.20
|
Uncharacterized protein |
| chr2_+_5832799 | 0.15 |
ENST00000322002.3
|
SOX11
|
SRY (sex determining region Y)-box 11 |
| chr4_-_140477910 | 0.15 |
ENST00000404104.3
|
SETD7
|
SET domain containing (lysine methyltransferase) 7 |
| chr4_+_15376165 | 0.15 |
ENST00000382383.3
ENST00000429690.1 |
C1QTNF7
|
C1q and tumor necrosis factor related protein 7 |
| chr2_-_202222091 | 0.15 |
ENST00000405148.2
ENST00000392257.3 ENST00000439709.1 |
ALS2CR12
|
amyotrophic lateral sclerosis 2 (juvenile) chromosome region, candidate 12 |
| chr5_+_102200948 | 0.15 |
ENST00000511477.1
ENST00000506006.1 ENST00000509832.1 |
PAM
|
peptidylglycine alpha-amidating monooxygenase |
| chrM_+_10464 | 0.15 |
ENST00000361335.1
|
MT-ND4L
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr8_+_96281045 | 0.15 |
ENST00000521905.1
|
KB-1047C11.2
|
KB-1047C11.2 |
| chr10_+_114710211 | 0.15 |
ENST00000349937.2
ENST00000369397.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr10_+_71561630 | 0.15 |
ENST00000398974.3
ENST00000398971.3 ENST00000398968.3 ENST00000398966.3 ENST00000398964.3 ENST00000398969.3 ENST00000356340.3 ENST00000398972.3 ENST00000398973.3 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr19_+_57751973 | 0.15 |
ENST00000535550.1
|
ZNF805
|
zinc finger protein 805 |
| chr20_-_44519839 | 0.15 |
ENST00000372518.4
|
NEURL2
|
neuralized E3 ubiquitin protein ligase 2 |
| chr9_+_129097520 | 0.15 |
ENST00000436593.3
|
MVB12B
|
multivesicular body subunit 12B |
| chr1_+_40862501 | 0.15 |
ENST00000539317.1
|
SMAP2
|
small ArfGAP2 |
| chr14_-_92414055 | 0.15 |
ENST00000342058.4
|
FBLN5
|
fibulin 5 |
| chr6_+_21666633 | 0.15 |
ENST00000606851.1
|
CASC15
|
cancer susceptibility candidate 15 (non-protein coding) |
| chr19_+_57752022 | 0.15 |
ENST00000354309.4
|
ZNF805
|
zinc finger protein 805 |
| chr13_+_28813645 | 0.15 |
ENST00000282391.5
|
PAN3
|
PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr4_+_155484103 | 0.15 |
ENST00000302068.4
|
FGB
|
fibrinogen beta chain |
| chr11_+_108535849 | 0.14 |
ENST00000526794.1
|
DDX10
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 10 |
| chr3_-_121379739 | 0.14 |
ENST00000428394.2
ENST00000314583.3 |
HCLS1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr8_-_108510224 | 0.14 |
ENST00000517746.1
ENST00000297450.3 |
ANGPT1
|
angiopoietin 1 |
| chr18_+_3466248 | 0.14 |
ENST00000581029.1
ENST00000581442.1 ENST00000579007.1 |
RP11-838N2.4
|
RP11-838N2.4 |
| chr10_-_105110831 | 0.14 |
ENST00000337211.4
|
PCGF6
|
polycomb group ring finger 6 |
| chr7_-_83824449 | 0.14 |
ENST00000420047.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr2_-_239140011 | 0.14 |
ENST00000409376.1
ENST00000409070.1 ENST00000409942.1 |
AC016757.3
|
Protein LOC151174 |
| chrX_-_84363974 | 0.14 |
ENST00000395409.3
ENST00000332921.5 ENST00000509231.1 |
SATL1
|
spermidine/spermine N1-acetyl transferase-like 1 |
| chr8_+_67104940 | 0.14 |
ENST00000517689.1
|
LINC00967
|
long intergenic non-protein coding RNA 967 |
| chr8_+_85095553 | 0.14 |
ENST00000521268.1
|
RALYL
|
RALY RNA binding protein-like |
| chr15_-_42749711 | 0.14 |
ENST00000565611.1
ENST00000263805.4 ENST00000565948.1 |
ZNF106
|
zinc finger protein 106 |
| chr10_-_61513146 | 0.14 |
ENST00000430431.1
|
LINC00948
|
long intergenic non-protein coding RNA 948 |
| chr12_+_28605426 | 0.14 |
ENST00000542801.1
|
CCDC91
|
coiled-coil domain containing 91 |
| chr5_-_127674883 | 0.14 |
ENST00000507835.1
|
FBN2
|
fibrillin 2 |
| chr17_+_67957878 | 0.14 |
ENST00000420427.1
|
AC004562.1
|
AC004562.1 |
| chr2_-_111291587 | 0.14 |
ENST00000437167.1
|
RGPD6
|
RANBP2-like and GRIP domain containing 6 |
| chr10_-_61122220 | 0.14 |
ENST00000422313.2
ENST00000435852.2 ENST00000442566.3 ENST00000373868.2 ENST00000277705.6 ENST00000373867.3 ENST00000419214.2 |
FAM13C
|
family with sequence similarity 13, member C |
| chr4_-_18023350 | 0.14 |
ENST00000539056.1
ENST00000382226.5 ENST00000326877.4 |
LCORL
|
ligand dependent nuclear receptor corepressor-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXD3_FOXI1_FOXF1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.1 | 0.4 | GO:0044029 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.1 | 0.4 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.1 | 0.3 | GO:2001035 | tongue muscle cell differentiation(GO:0035981) positive regulation of skeletal muscle fiber differentiation(GO:1902811) regulation of tongue muscle cell differentiation(GO:2001035) positive regulation of tongue muscle cell differentiation(GO:2001037) |
| 0.1 | 0.4 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.1 | 0.4 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.1 | 0.3 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.1 | 0.2 | GO:0060730 | regulation of intestinal epithelial structure maintenance(GO:0060730) |
| 0.1 | 0.3 | GO:1904823 | pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.1 | 0.2 | GO:1903676 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.1 | 0.5 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.1 | 0.2 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.1 | 0.5 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.1 | 0.2 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 0.2 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.4 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.0 | 0.3 | GO:0043606 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.3 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.0 | 0.3 | GO:1903786 | mycotoxin catabolic process(GO:0043387) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound catabolic process(GO:1901377) regulation of glutathione biosynthetic process(GO:1903786) positive regulation of glutathione biosynthetic process(GO:1903788) |
| 0.0 | 0.1 | GO:0032918 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.0 | 0.3 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.4 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.1 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.0 | 0.2 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.5 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.2 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.1 | GO:0001897 | cytolysis by symbiont of host cells(GO:0001897) |
| 0.0 | 0.2 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.3 | GO:0021590 | cerebellum maturation(GO:0021590) cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.2 | GO:2000490 | negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.0 | 0.3 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 | 0.1 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 0.4 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.1 | GO:2000705 | dense core granule biogenesis(GO:0061110) regulation of dense core granule biogenesis(GO:2000705) |
| 0.0 | 0.2 | GO:0072096 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.0 | 1.0 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.0 | 0.4 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.1 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.0 | 0.2 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.2 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.0 | GO:0035378 | carbon dioxide transmembrane transport(GO:0035378) |
| 0.0 | 0.1 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.2 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.1 | GO:0060940 | epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) |
| 0.0 | 0.2 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.1 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.0 | 0.4 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.8 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.3 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.0 | 0.1 | GO:0045763 | negative regulation of cellular amino acid metabolic process(GO:0045763) |
| 0.0 | 0.1 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.0 | 0.2 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.0 | 0.0 | GO:0045410 | positive regulation of interleukin-6 biosynthetic process(GO:0045410) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:1901896 | positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.0 | 0.1 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.0 | 0.2 | GO:0018032 | protein amidation(GO:0018032) |
| 0.0 | 0.1 | GO:0045720 | negative regulation of integrin biosynthetic process(GO:0045720) |
| 0.0 | 0.2 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.0 | 0.1 | GO:0033385 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.0 | 0.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.2 | GO:0061370 | testosterone biosynthetic process(GO:0061370) |
| 0.0 | 0.2 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.1 | GO:0060151 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.0 | 0.1 | GO:0051622 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.0 | 0.8 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 0.3 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 0.1 | GO:0002522 | leukocyte migration involved in immune response(GO:0002522) |
| 0.0 | 0.1 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.0 | 0.1 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.0 | 0.1 | GO:0045354 | negative regulation of interferon-alpha production(GO:0032687) interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) |
| 0.0 | 0.2 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.0 | 0.1 | GO:0019075 | virus maturation(GO:0019075) |
| 0.0 | 0.0 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.0 | 0.1 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.0 | 0.2 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.2 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.5 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.1 | GO:2000426 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) negative regulation of apoptotic cell clearance(GO:2000426) |
| 0.0 | 0.1 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.0 | 0.2 | GO:0035766 | cell chemotaxis to fibroblast growth factor(GO:0035766) endothelial cell chemotaxis to fibroblast growth factor(GO:0035768) regulation of cell chemotaxis to fibroblast growth factor(GO:1904847) regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000544) |
| 0.0 | 0.1 | GO:0021764 | amygdala development(GO:0021764) |
| 0.0 | 0.1 | GO:0009258 | 10-formyltetrahydrofolate catabolic process(GO:0009258) |
| 0.0 | 0.2 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.0 | GO:0007623 | circadian rhythm(GO:0007623) |
| 0.0 | 0.1 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.0 | 0.1 | GO:0002760 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.0 | 0.5 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.1 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.0 | 0.1 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 0.2 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.1 | GO:0097534 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.0 | 0.1 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 0.0 | 0.1 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.0 | 0.1 | GO:0007210 | serotonin receptor signaling pathway(GO:0007210) |
| 0.0 | 0.3 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.1 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.0 | 0.3 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.1 | GO:0006286 | base-excision repair, base-free sugar-phosphate removal(GO:0006286) |
| 0.0 | 0.2 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.0 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.0 | 0.0 | GO:0021943 | formation of radial glial scaffolds(GO:0021943) |
| 0.0 | 0.0 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.1 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.1 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.1 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) actin filament branching(GO:0090135) |
| 0.0 | 0.1 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.0 | 0.0 | GO:0060738 | epithelial-mesenchymal signaling involved in prostate gland development(GO:0060738) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.0 | GO:0018879 | biphenyl metabolic process(GO:0018879) |
| 0.0 | 0.2 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 0.1 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.2 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.1 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.0 | 0.0 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.1 | GO:0044793 | negative regulation by host of viral process(GO:0044793) |
| 0.0 | 0.1 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.0 | GO:1903926 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.0 | 0.0 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.0 | 0.5 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.2 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.0 | 0.2 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.3 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.5 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.2 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.3 | GO:0033630 | positive regulation of cell adhesion mediated by integrin(GO:0033630) |
| 0.0 | 0.1 | GO:0018022 | peptidyl-lysine methylation(GO:0018022) |
| 0.0 | 0.9 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.2 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.0 | 0.1 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.3 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.0 | GO:1902990 | leading strand elongation(GO:0006272) mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.0 | 0.2 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.1 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.1 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.1 | GO:0010166 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.0 | 0.1 | GO:0007269 | neurotransmitter secretion(GO:0007269) signal release from synapse(GO:0099643) |
| 0.0 | 0.1 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.0 | 0.0 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.0 | 0.1 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.1 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 0.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.0 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 | 0.0 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.0 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.1 | 1.0 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.1 | 0.3 | GO:0097232 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.1 | 0.2 | GO:0005879 | axonemal microtubule(GO:0005879) |
| 0.1 | 0.4 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.1 | 0.2 | GO:0030936 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.1 | 0.2 | GO:0043614 | multi-eIF complex(GO:0043614) translation preinitiation complex(GO:0070993) astrocyte end-foot(GO:0097450) |
| 0.1 | 0.4 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 0.2 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.8 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.2 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.4 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.1 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.0 | 0.6 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.5 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0098576 | lumenal side of membrane(GO:0098576) |
| 0.0 | 0.1 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.1 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 0.2 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.1 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.0 | 0.8 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.2 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 0.0 | 0.1 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.1 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.0 | 0.2 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.0 | 0.0 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.1 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 0.1 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.1 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.0 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.0 | 0.1 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.2 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.1 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.0 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.4 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.8 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.2 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.1 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.4 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.0 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 0.0 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.2 | GO:0033391 | chromatoid body(GO:0033391) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 0.1 | 0.5 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.1 | 0.3 | GO:0004397 | histidine ammonia-lyase activity(GO:0004397) |
| 0.1 | 0.4 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.1 | 0.2 | GO:0031862 | prostanoid receptor binding(GO:0031862) |
| 0.1 | 0.8 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.1 | 0.3 | GO:0015265 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) glycerol channel activity(GO:0015254) urea channel activity(GO:0015265) |
| 0.1 | 0.8 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.2 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 0.1 | 0.2 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.1 | 0.3 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.1 | 0.4 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.1 | 0.3 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) polyamine binding(GO:0019808) |
| 0.0 | 0.3 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.2 | GO:0001228 | transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.1 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.2 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.0 | 0.2 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 0.0 | 0.2 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.0 | 0.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.2 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.2 | GO:0050119 | N-acetylglucosamine deacetylase activity(GO:0050119) |
| 0.0 | 0.3 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.0 | 0.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.2 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.3 | GO:0004883 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.2 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.1 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.0 | 0.1 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.2 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.0 | 0.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0070260 | tyrosyl-RNA phosphodiesterase activity(GO:0036317) 5'-tyrosyl-DNA phosphodiesterase activity(GO:0070260) |
| 0.0 | 0.1 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.0 | 0.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.0 | 0.0 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.0 | 0.4 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.4 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.1 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.0 | 0.3 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.2 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.1 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.0 | 0.2 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.1 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 0.1 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.2 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.0 | 0.4 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.0 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.0 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 0.0 | 0.4 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.1 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 0.1 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.0 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.1 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.1 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 0.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.1 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 0.0 | 0.2 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 0.1 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0004057 | arginyltransferase activity(GO:0004057) |
| 0.0 | 0.0 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.9 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) steroid sulfotransferase activity(GO:0050294) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.2 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.0 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.1 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 1.7 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.0 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.0 | 0.1 | GO:0044388 | small protein activating enzyme binding(GO:0044388) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.1 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.2 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.1 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.0 | 0.1 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.1 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.2 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.4 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.2 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 0.2 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 0.1 | GO:1902444 | riboflavin binding(GO:1902444) |
| 0.0 | 0.1 | GO:0015185 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.6 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.7 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.1 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.5 | PID MYC PATHWAY | C-MYC pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.5 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.3 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 1.5 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.4 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.4 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.8 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.6 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.4 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.2 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.1 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |