Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
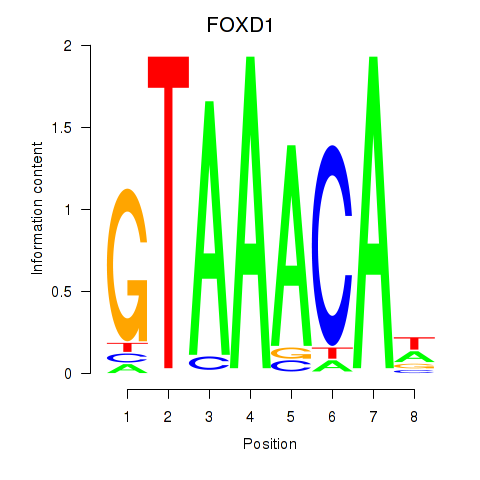
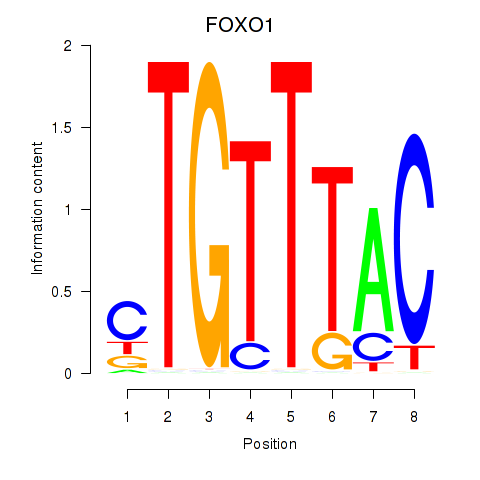
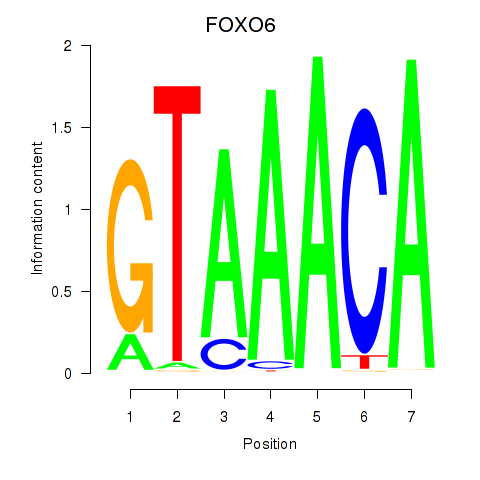
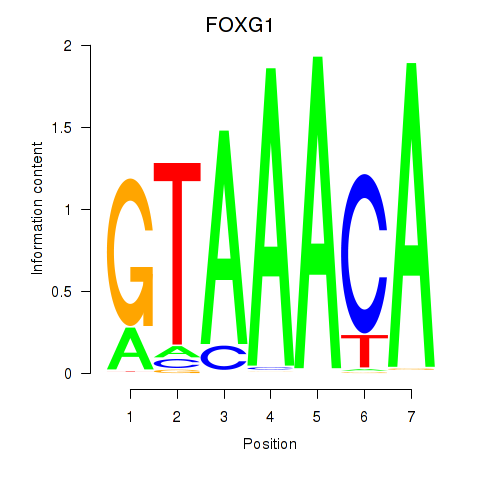
Results for FOXD1_FOXO1_FOXO6_FOXG1_FOXP1
Z-value: 1.00
Transcription factors associated with FOXD1_FOXO1_FOXO6_FOXG1_FOXP1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXD1
|
ENSG00000251493.2 | forkhead box D1 |
|
FOXO1
|
ENSG00000150907.6 | forkhead box O1 |
|
FOXO6
|
ENSG00000204060.4 | forkhead box O6 |
|
FOXG1
|
ENSG00000176165.7 | forkhead box G1 |
|
FOXP1
|
ENSG00000114861.14 | forkhead box P1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXP1 | hg19_v2_chr3_-_71179988_71180085 | 0.98 | 1.7e-02 | Click! |
| FOXD1 | hg19_v2_chr5_-_72744336_72744359 | 0.95 | 5.4e-02 | Click! |
| FOXO6 | hg19_v2_chr1_+_41827594_41827594 | -0.56 | 4.4e-01 | Click! |
| FOXG1 | hg19_v2_chr14_+_29236269_29236287 | -0.53 | 4.7e-01 | Click! |
| FOXO1 | hg19_v2_chr13_-_41240717_41240735 | -0.30 | 7.0e-01 | Click! |
Activity profile of FOXD1_FOXO1_FOXO6_FOXG1_FOXP1 motif
Sorted Z-values of FOXD1_FOXO1_FOXO6_FOXG1_FOXP1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_84563295 | 1.53 |
ENST00000369687.1
|
RIPPLY2
|
ripply transcriptional repressor 2 |
| chr2_+_66918558 | 1.51 |
ENST00000435389.1
ENST00000428590.1 ENST00000412944.1 |
AC007392.3
|
AC007392.3 |
| chr7_+_134832808 | 1.26 |
ENST00000275767.3
|
TMEM140
|
transmembrane protein 140 |
| chr2_-_191878681 | 1.11 |
ENST00000409465.1
|
STAT1
|
signal transducer and activator of transcription 1, 91kDa |
| chr1_+_116654376 | 1.08 |
ENST00000369500.3
|
MAB21L3
|
mab-21-like 3 (C. elegans) |
| chr17_+_26662679 | 1.02 |
ENST00000578158.1
|
TNFAIP1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chr12_-_116714564 | 0.94 |
ENST00000548743.1
|
MED13L
|
mediator complex subunit 13-like |
| chr1_-_161337662 | 0.91 |
ENST00000367974.1
|
C1orf192
|
chromosome 1 open reading frame 192 |
| chr18_-_2982869 | 0.91 |
ENST00000584915.1
|
LPIN2
|
lipin 2 |
| chr22_-_31688431 | 0.87 |
ENST00000402249.3
ENST00000443175.1 ENST00000215912.5 ENST00000441972.1 |
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr10_+_43932282 | 0.81 |
ENST00000431662.1
ENST00000315429.6 |
ZNF487
|
zinc finger protein 487 |
| chr2_+_149402989 | 0.79 |
ENST00000397424.2
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr10_+_75668916 | 0.79 |
ENST00000481390.1
|
PLAU
|
plasminogen activator, urokinase |
| chr4_-_141075330 | 0.77 |
ENST00000509479.2
|
MAML3
|
mastermind-like 3 (Drosophila) |
| chr8_-_29208183 | 0.76 |
ENST00000240100.2
|
DUSP4
|
dual specificity phosphatase 4 |
| chr5_-_150460539 | 0.74 |
ENST00000520931.1
ENST00000520695.1 ENST00000521591.1 ENST00000518977.1 |
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr7_-_112579673 | 0.74 |
ENST00000432572.1
|
C7orf60
|
chromosome 7 open reading frame 60 |
| chr3_-_185826718 | 0.74 |
ENST00000413301.1
ENST00000421809.1 |
ETV5
|
ets variant 5 |
| chr3_-_71179988 | 0.73 |
ENST00000491238.1
|
FOXP1
|
forkhead box P1 |
| chr12_+_13349711 | 0.73 |
ENST00000538364.1
ENST00000396301.3 |
EMP1
|
epithelial membrane protein 1 |
| chr6_+_26020672 | 0.73 |
ENST00000357647.3
|
HIST1H3A
|
histone cluster 1, H3a |
| chr13_-_46716969 | 0.69 |
ENST00000435666.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr6_-_88875628 | 0.67 |
ENST00000551417.1
|
CNR1
|
cannabinoid receptor 1 (brain) |
| chr5_+_133451254 | 0.66 |
ENST00000517851.1
ENST00000521639.1 ENST00000522375.1 ENST00000378560.4 ENST00000432532.2 ENST00000520958.1 ENST00000518915.1 ENST00000395023.1 |
TCF7
|
transcription factor 7 (T-cell specific, HMG-box) |
| chr17_+_72427477 | 0.63 |
ENST00000342648.5
ENST00000481232.1 |
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr8_+_95908041 | 0.63 |
ENST00000396113.1
ENST00000519136.1 |
NDUFAF6
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr8_+_39972170 | 0.62 |
ENST00000521257.1
|
RP11-359E19.2
|
RP11-359E19.2 |
| chr17_+_65373531 | 0.61 |
ENST00000580974.1
|
PITPNC1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr7_-_99698338 | 0.61 |
ENST00000354230.3
ENST00000425308.1 |
MCM7
|
minichromosome maintenance complex component 7 |
| chr2_+_27665232 | 0.60 |
ENST00000543753.1
ENST00000288873.3 |
KRTCAP3
|
keratinocyte associated protein 3 |
| chr22_-_31688381 | 0.59 |
ENST00000487265.2
|
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr3_-_185826855 | 0.58 |
ENST00000306376.5
|
ETV5
|
ets variant 5 |
| chr15_+_96897466 | 0.58 |
ENST00000558382.1
ENST00000558499.1 |
RP11-522B15.3
|
RP11-522B15.3 |
| chr5_-_137674000 | 0.57 |
ENST00000510119.1
ENST00000513970.1 |
CDC25C
|
cell division cycle 25C |
| chr17_+_72426891 | 0.56 |
ENST00000392627.1
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr18_-_59415987 | 0.56 |
ENST00000590199.1
ENST00000590968.1 |
RP11-879F14.1
|
RP11-879F14.1 |
| chr1_+_66796401 | 0.55 |
ENST00000528771.1
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr16_-_84150392 | 0.55 |
ENST00000570012.1
|
MBTPS1
|
membrane-bound transcription factor peptidase, site 1 |
| chr2_-_165424973 | 0.53 |
ENST00000543549.1
|
GRB14
|
growth factor receptor-bound protein 14 |
| chr5_-_150460914 | 0.52 |
ENST00000389378.2
|
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr1_+_92545862 | 0.51 |
ENST00000370382.3
ENST00000342818.3 |
BTBD8
|
BTB (POZ) domain containing 8 |
| chr17_+_37821593 | 0.50 |
ENST00000578283.1
|
TCAP
|
titin-cap |
| chr2_+_33661382 | 0.50 |
ENST00000402538.3
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr12_+_13349650 | 0.49 |
ENST00000256951.5
ENST00000431267.2 ENST00000542474.1 ENST00000544053.1 |
EMP1
|
epithelial membrane protein 1 |
| chr16_-_57832004 | 0.49 |
ENST00000562503.1
|
KIFC3
|
kinesin family member C3 |
| chr1_-_12677714 | 0.49 |
ENST00000376223.2
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr1_-_27816556 | 0.49 |
ENST00000536657.1
|
WASF2
|
WAS protein family, member 2 |
| chr17_+_65374075 | 0.48 |
ENST00000581322.1
|
PITPNC1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr7_+_129007964 | 0.48 |
ENST00000460109.1
ENST00000474594.1 ENST00000446212.1 |
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr2_+_175260514 | 0.48 |
ENST00000424069.1
ENST00000427038.1 |
SCRN3
|
secernin 3 |
| chr10_+_97759848 | 0.47 |
ENST00000424464.1
ENST00000410012.2 ENST00000344386.3 |
CC2D2B
|
coiled-coil and C2 domain containing 2B |
| chr2_+_149402553 | 0.47 |
ENST00000258484.6
ENST00000409654.1 |
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr1_+_209878182 | 0.47 |
ENST00000367027.3
|
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1 |
| chrX_+_135251835 | 0.47 |
ENST00000456445.1
|
FHL1
|
four and a half LIM domains 1 |
| chr3_-_117716418 | 0.46 |
ENST00000484092.1
|
RP11-384F7.2
|
RP11-384F7.2 |
| chr11_+_10476851 | 0.45 |
ENST00000396553.2
|
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr9_-_20382446 | 0.44 |
ENST00000380321.1
|
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr1_+_152178320 | 0.44 |
ENST00000429352.1
|
RP11-107M16.2
|
RP11-107M16.2 |
| chr9_-_84303269 | 0.44 |
ENST00000418319.1
|
TLE1
|
transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila) |
| chrX_+_135251783 | 0.44 |
ENST00000394153.2
|
FHL1
|
four and a half LIM domains 1 |
| chr17_+_41561317 | 0.43 |
ENST00000540306.1
ENST00000262415.3 ENST00000605777.1 |
DHX8
|
DEAH (Asp-Glu-Ala-His) box polypeptide 8 |
| chr2_+_27665289 | 0.43 |
ENST00000407293.1
|
KRTCAP3
|
keratinocyte associated protein 3 |
| chr2_-_160472952 | 0.41 |
ENST00000541068.2
ENST00000355831.2 ENST00000343439.5 ENST00000392782.1 |
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B |
| chr11_+_117049910 | 0.41 |
ENST00000431081.2
ENST00000524842.1 |
SIDT2
|
SID1 transmembrane family, member 2 |
| chr2_-_176033066 | 0.41 |
ENST00000437522.1
|
ATF2
|
activating transcription factor 2 |
| chr19_+_41725140 | 0.41 |
ENST00000359092.3
|
AXL
|
AXL receptor tyrosine kinase |
| chr20_-_45984401 | 0.40 |
ENST00000311275.7
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr11_-_115375107 | 0.39 |
ENST00000545380.1
ENST00000452722.3 ENST00000537058.1 ENST00000536727.1 ENST00000542447.2 ENST00000331581.6 |
CADM1
|
cell adhesion molecule 1 |
| chrX_+_135252050 | 0.39 |
ENST00000449474.1
ENST00000345434.3 |
FHL1
|
four and a half LIM domains 1 |
| chr14_+_24584508 | 0.39 |
ENST00000559354.1
ENST00000560459.1 ENST00000559593.1 ENST00000396941.4 ENST00000396936.1 |
DCAF11
|
DDB1 and CUL4 associated factor 11 |
| chr12_+_54378849 | 0.39 |
ENST00000515593.1
|
HOXC10
|
homeobox C10 |
| chr5_-_16742330 | 0.39 |
ENST00000505695.1
ENST00000427430.2 |
MYO10
|
myosin X |
| chr1_+_3541543 | 0.38 |
ENST00000378344.2
ENST00000344579.5 |
TPRG1L
|
tumor protein p63 regulated 1-like |
| chr8_-_119124045 | 0.38 |
ENST00000378204.2
|
EXT1
|
exostosin glycosyltransferase 1 |
| chr2_-_27434611 | 0.38 |
ENST00000408041.1
|
SLC5A6
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
| chr12_+_48499252 | 0.38 |
ENST00000549003.1
ENST00000550924.1 |
PFKM
|
phosphofructokinase, muscle |
| chr3_-_48956818 | 0.37 |
ENST00000408959.2
|
ARIH2OS
|
ariadne homolog 2 opposite strand |
| chr19_-_45909585 | 0.36 |
ENST00000593226.1
ENST00000418234.2 |
PPP1R13L
|
protein phosphatase 1, regulatory subunit 13 like |
| chr2_+_198380763 | 0.36 |
ENST00000448447.2
ENST00000409360.1 |
MOB4
|
MOB family member 4, phocein |
| chr17_+_26662597 | 0.36 |
ENST00000544907.2
|
TNFAIP1
|
tumor necrosis factor, alpha-induced protein 1 (endothelial) |
| chr2_-_233877912 | 0.36 |
ENST00000264051.3
|
NGEF
|
neuronal guanine nucleotide exchange factor |
| chr11_+_31833939 | 0.35 |
ENST00000530348.1
|
RCN1
|
reticulocalbin 1, EF-hand calcium binding domain |
| chr6_-_30043539 | 0.35 |
ENST00000376751.3
ENST00000244360.6 |
RNF39
|
ring finger protein 39 |
| chr3_-_178865747 | 0.34 |
ENST00000435560.1
|
RP11-360P21.2
|
RP11-360P21.2 |
| chr8_+_126442563 | 0.34 |
ENST00000311922.3
|
TRIB1
|
tribbles pseudokinase 1 |
| chr8_+_31496809 | 0.34 |
ENST00000518104.1
ENST00000519301.1 |
NRG1
|
neuregulin 1 |
| chr19_+_13906250 | 0.34 |
ENST00000254323.2
|
ZSWIM4
|
zinc finger, SWIM-type containing 4 |
| chr20_-_43150601 | 0.33 |
ENST00000541235.1
ENST00000255175.1 ENST00000342374.4 |
SERINC3
|
serine incorporator 3 |
| chr3_+_148447887 | 0.33 |
ENST00000475347.1
ENST00000474935.1 ENST00000461609.1 |
AGTR1
|
angiotensin II receptor, type 1 |
| chr10_+_31610064 | 0.33 |
ENST00000446923.2
ENST00000559476.1 |
ZEB1
|
zinc finger E-box binding homeobox 1 |
| chr11_+_3968573 | 0.33 |
ENST00000532990.1
|
STIM1
|
stromal interaction molecule 1 |
| chr5_-_115872142 | 0.32 |
ENST00000510263.1
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr7_-_25702669 | 0.31 |
ENST00000446840.1
|
AC003090.1
|
AC003090.1 |
| chr8_+_97773202 | 0.31 |
ENST00000519484.1
|
CPQ
|
carboxypeptidase Q |
| chr6_+_33168189 | 0.31 |
ENST00000444757.1
|
SLC39A7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr1_-_153931052 | 0.31 |
ENST00000368630.3
ENST00000368633.1 |
CRTC2
|
CREB regulated transcription coactivator 2 |
| chr16_-_57831676 | 0.31 |
ENST00000465878.2
ENST00000539578.1 ENST00000561524.1 |
KIFC3
|
kinesin family member C3 |
| chr17_-_1418972 | 0.30 |
ENST00000571274.1
|
INPP5K
|
inositol polyphosphate-5-phosphatase K |
| chr14_-_36988882 | 0.30 |
ENST00000498187.2
|
NKX2-1
|
NK2 homeobox 1 |
| chr16_+_57680840 | 0.30 |
ENST00000563862.1
ENST00000564722.1 ENST00000569158.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr14_-_37051798 | 0.30 |
ENST00000258829.5
|
NKX2-8
|
NK2 homeobox 8 |
| chr2_+_12857015 | 0.30 |
ENST00000155926.4
|
TRIB2
|
tribbles pseudokinase 2 |
| chr3_-_107777208 | 0.30 |
ENST00000398258.3
|
CD47
|
CD47 molecule |
| chr10_+_99400443 | 0.30 |
ENST00000370631.3
|
PI4K2A
|
phosphatidylinositol 4-kinase type 2 alpha |
| chr1_+_43855545 | 0.29 |
ENST00000372450.4
ENST00000310739.4 |
SZT2
|
seizure threshold 2 homolog (mouse) |
| chr16_-_57831914 | 0.29 |
ENST00000421376.2
|
KIFC3
|
kinesin family member C3 |
| chr2_-_74007095 | 0.28 |
ENST00000452812.1
|
DUSP11
|
dual specificity phosphatase 11 (RNA/RNP complex 1-interacting) |
| chr14_+_23067166 | 0.28 |
ENST00000216327.6
ENST00000542041.1 |
ABHD4
|
abhydrolase domain containing 4 |
| chr6_-_134638767 | 0.28 |
ENST00000524929.1
|
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr17_+_9745786 | 0.28 |
ENST00000304773.5
|
GLP2R
|
glucagon-like peptide 2 receptor |
| chr1_-_27816641 | 0.28 |
ENST00000430629.2
|
WASF2
|
WAS protein family, member 2 |
| chr7_+_134576317 | 0.27 |
ENST00000424922.1
ENST00000495522.1 |
CALD1
|
caldesmon 1 |
| chr2_-_152146385 | 0.27 |
ENST00000414946.1
ENST00000243346.5 |
NMI
|
N-myc (and STAT) interactor |
| chr8_+_97773457 | 0.27 |
ENST00000521142.1
|
CPQ
|
carboxypeptidase Q |
| chr9_-_135819987 | 0.27 |
ENST00000298552.3
ENST00000403810.1 |
TSC1
|
tuberous sclerosis 1 |
| chr16_+_86612112 | 0.27 |
ENST00000320241.3
|
FOXL1
|
forkhead box L1 |
| chr1_+_164528437 | 0.26 |
ENST00000485769.1
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr4_-_41884620 | 0.26 |
ENST00000504870.1
|
LINC00682
|
long intergenic non-protein coding RNA 682 |
| chr9_+_131904233 | 0.26 |
ENST00000432651.1
ENST00000435132.1 |
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr1_-_28503693 | 0.26 |
ENST00000373857.3
|
PTAFR
|
platelet-activating factor receptor |
| chr12_+_120740119 | 0.26 |
ENST00000536460.1
ENST00000202967.4 |
SIRT4
|
sirtuin 4 |
| chr2_-_27434635 | 0.26 |
ENST00000401463.1
|
SLC5A6
|
solute carrier family 5 (sodium/multivitamin and iodide cotransporter), member 6 |
| chr9_-_16253112 | 0.26 |
ENST00000380683.1
|
C9orf92
|
chromosome 9 open reading frame 92 |
| chr1_+_84630574 | 0.26 |
ENST00000413538.1
ENST00000417530.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr4_-_69215699 | 0.25 |
ENST00000510746.1
ENST00000344157.4 ENST00000355665.3 |
YTHDC1
|
YTH domain containing 1 |
| chr15_+_80351910 | 0.25 |
ENST00000261749.6
ENST00000561060.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr9_+_131903916 | 0.25 |
ENST00000419582.1
|
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr18_-_53303123 | 0.25 |
ENST00000569357.1
ENST00000565124.1 ENST00000398339.1 |
TCF4
|
transcription factor 4 |
| chr1_+_20512568 | 0.25 |
ENST00000375099.3
|
UBXN10
|
UBX domain protein 10 |
| chr10_-_101380121 | 0.25 |
ENST00000370495.4
|
SLC25A28
|
solute carrier family 25 (mitochondrial iron transporter), member 28 |
| chr3_-_71179699 | 0.25 |
ENST00000497355.1
|
FOXP1
|
forkhead box P1 |
| chr3_-_71294304 | 0.24 |
ENST00000498215.1
|
FOXP1
|
forkhead box P1 |
| chr16_+_57679859 | 0.24 |
ENST00000569494.1
ENST00000566169.1 |
GPR56
|
G protein-coupled receptor 56 |
| chr15_-_83240507 | 0.24 |
ENST00000564522.1
ENST00000398592.2 |
CPEB1
|
cytoplasmic polyadenylation element binding protein 1 |
| chr2_-_190927447 | 0.24 |
ENST00000260950.4
|
MSTN
|
myostatin |
| chrX_-_106146547 | 0.24 |
ENST00000276173.4
ENST00000411805.1 |
RIPPLY1
|
ripply transcriptional repressor 1 |
| chr3_+_107241783 | 0.24 |
ENST00000415149.2
ENST00000402543.1 ENST00000325805.8 ENST00000427402.1 |
BBX
|
bobby sox homolog (Drosophila) |
| chr6_-_42418999 | 0.24 |
ENST00000340840.2
ENST00000354325.2 |
TRERF1
|
transcriptional regulating factor 1 |
| chrX_-_54070607 | 0.24 |
ENST00000338154.6
ENST00000338946.6 |
PHF8
|
PHD finger protein 8 |
| chr1_+_179851893 | 0.24 |
ENST00000531630.2
|
TOR1AIP1
|
torsin A interacting protein 1 |
| chr15_+_80733570 | 0.23 |
ENST00000533983.1
ENST00000527771.1 ENST00000525103.1 |
ARNT2
|
aryl-hydrocarbon receptor nuclear translocator 2 |
| chr11_-_34535297 | 0.23 |
ENST00000532417.1
|
ELF5
|
E74-like factor 5 (ets domain transcription factor) |
| chr6_-_127780510 | 0.22 |
ENST00000487331.2
ENST00000483725.3 |
KIAA0408
|
KIAA0408 |
| chr16_+_57680811 | 0.22 |
ENST00000569101.1
|
GPR56
|
G protein-coupled receptor 56 |
| chr6_+_30853002 | 0.22 |
ENST00000421124.2
ENST00000512725.1 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr13_-_46756351 | 0.22 |
ENST00000323076.2
|
LCP1
|
lymphocyte cytosolic protein 1 (L-plastin) |
| chr9_+_131904295 | 0.22 |
ENST00000434095.1
|
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr2_+_12857043 | 0.22 |
ENST00000381465.2
|
TRIB2
|
tribbles pseudokinase 2 |
| chr7_-_27219849 | 0.22 |
ENST00000396344.4
|
HOXA10
|
homeobox A10 |
| chr16_-_69418553 | 0.22 |
ENST00000569542.2
|
TERF2
|
telomeric repeat binding factor 2 |
| chr6_+_15401075 | 0.22 |
ENST00000541660.1
|
JARID2
|
jumonji, AT rich interactive domain 2 |
| chr3_+_171561127 | 0.22 |
ENST00000334567.5
ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr8_-_133772794 | 0.22 |
ENST00000519187.1
ENST00000523829.1 ENST00000356838.3 ENST00000377901.4 ENST00000519304.1 |
TMEM71
|
transmembrane protein 71 |
| chr20_+_44421137 | 0.22 |
ENST00000415790.1
|
DNTTIP1
|
deoxynucleotidyltransferase, terminal, interacting protein 1 |
| chr6_-_33168391 | 0.21 |
ENST00000374685.4
ENST00000413614.2 ENST00000374680.3 |
RXRB
|
retinoid X receptor, beta |
| chr16_+_57679945 | 0.21 |
ENST00000568157.1
|
GPR56
|
G protein-coupled receptor 56 |
| chr8_+_81397846 | 0.21 |
ENST00000379091.4
|
ZBTB10
|
zinc finger and BTB domain containing 10 |
| chr6_+_32121908 | 0.21 |
ENST00000375143.2
ENST00000424499.1 |
PPT2
|
palmitoyl-protein thioesterase 2 |
| chr19_+_41725088 | 0.21 |
ENST00000301178.4
|
AXL
|
AXL receptor tyrosine kinase |
| chrX_-_19688475 | 0.21 |
ENST00000541422.1
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chrX_-_15619076 | 0.21 |
ENST00000252519.3
|
ACE2
|
angiotensin I converting enzyme 2 |
| chr1_-_168106536 | 0.21 |
ENST00000537209.1
ENST00000361697.2 ENST00000546300.1 ENST00000367835.1 |
GPR161
|
G protein-coupled receptor 161 |
| chr11_+_117049854 | 0.21 |
ENST00000278951.7
|
SIDT2
|
SID1 transmembrane family, member 2 |
| chr11_+_63606477 | 0.20 |
ENST00000508192.1
ENST00000361128.5 |
MARK2
|
MAP/microtubule affinity-regulating kinase 2 |
| chr2_-_68547061 | 0.20 |
ENST00000263655.3
|
CNRIP1
|
cannabinoid receptor interacting protein 1 |
| chr20_+_44035200 | 0.20 |
ENST00000372717.1
ENST00000360981.4 |
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr11_+_46402297 | 0.20 |
ENST00000405308.2
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chrX_-_68385354 | 0.20 |
ENST00000361478.1
|
PJA1
|
praja ring finger 1, E3 ubiquitin protein ligase |
| chr2_-_26205550 | 0.20 |
ENST00000405914.1
|
KIF3C
|
kinesin family member 3C |
| chr12_-_6665200 | 0.20 |
ENST00000336604.4
ENST00000396840.2 ENST00000356896.4 |
IFFO1
|
intermediate filament family orphan 1 |
| chr3_+_137717571 | 0.20 |
ENST00000343735.4
|
CLDN18
|
claudin 18 |
| chr16_-_69418649 | 0.20 |
ENST00000566257.1
|
TERF2
|
telomeric repeat binding factor 2 |
| chr5_+_121465207 | 0.20 |
ENST00000296600.4
|
ZNF474
|
zinc finger protein 474 |
| chr20_+_56964169 | 0.20 |
ENST00000475243.1
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr9_-_107690420 | 0.20 |
ENST00000423487.2
ENST00000374733.1 ENST00000374736.3 |
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1 |
| chr2_+_70142232 | 0.20 |
ENST00000540449.1
|
MXD1
|
MAX dimerization protein 1 |
| chr3_-_71632894 | 0.19 |
ENST00000493089.1
|
FOXP1
|
forkhead box P1 |
| chr10_-_13570533 | 0.19 |
ENST00000396900.2
ENST00000396898.2 |
BEND7
|
BEN domain containing 7 |
| chr22_-_45404819 | 0.19 |
ENST00000447824.3
ENST00000404079.2 ENST00000420689.1 ENST00000403565.1 |
PHF21B
|
PHD finger protein 21B |
| chr2_-_26205340 | 0.19 |
ENST00000264712.3
|
KIF3C
|
kinesin family member 3C |
| chr11_+_46402482 | 0.19 |
ENST00000441869.1
|
MDK
|
midkine (neurite growth-promoting factor 2) |
| chr17_+_74372662 | 0.19 |
ENST00000591651.1
ENST00000545180.1 |
SPHK1
|
sphingosine kinase 1 |
| chr17_-_39183452 | 0.19 |
ENST00000361883.5
|
KRTAP1-5
|
keratin associated protein 1-5 |
| chr13_+_50589390 | 0.19 |
ENST00000360473.4
ENST00000312942.1 |
KCNRG
|
potassium channel regulator |
| chr4_+_86396321 | 0.19 |
ENST00000503995.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr3_-_141868357 | 0.18 |
ENST00000489671.1
ENST00000475734.1 ENST00000467072.1 ENST00000499676.2 |
TFDP2
|
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr16_-_67597789 | 0.18 |
ENST00000605277.1
|
CTD-2012K14.6
|
CTD-2012K14.6 |
| chr16_-_87729753 | 0.18 |
ENST00000538868.1
|
AC010536.1
|
Uncharacterized protein; cDNA FLJ45526 fis, clone BRTHA2027227 |
| chr13_-_30683005 | 0.18 |
ENST00000413591.1
ENST00000432770.1 |
LINC00365
|
long intergenic non-protein coding RNA 365 |
| chr19_+_13134772 | 0.18 |
ENST00000587760.1
ENST00000585575.1 |
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr8_-_134309823 | 0.18 |
ENST00000414097.2
|
NDRG1
|
N-myc downstream regulated 1 |
| chr17_+_38333263 | 0.18 |
ENST00000456989.2
ENST00000543876.1 ENST00000544503.1 ENST00000264644.6 ENST00000538884.1 |
RAPGEFL1
|
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr1_-_94079648 | 0.18 |
ENST00000370247.3
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr12_-_12714006 | 0.18 |
ENST00000541207.1
|
DUSP16
|
dual specificity phosphatase 16 |
| chr10_-_46089939 | 0.18 |
ENST00000453980.3
|
MARCH8
|
membrane-associated ring finger (C3HC4) 8, E3 ubiquitin protein ligase |
| chr17_-_61776522 | 0.18 |
ENST00000582055.1
|
LIMD2
|
LIM domain containing 2 |
| chr8_-_134309335 | 0.18 |
ENST00000522890.1
ENST00000323851.7 ENST00000518176.1 ENST00000354944.5 ENST00000537882.1 ENST00000522476.1 ENST00000518066.1 ENST00000521544.1 ENST00000518480.1 ENST00000523892.1 |
NDRG1
|
N-myc downstream regulated 1 |
| chr12_+_50135351 | 0.18 |
ENST00000549445.1
ENST00000550951.1 ENST00000549385.1 ENST00000548713.1 ENST00000548201.1 |
TMBIM6
|
transmembrane BAX inhibitor motif containing 6 |
| chr17_+_72428218 | 0.18 |
ENST00000392628.2
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr15_-_34629922 | 0.18 |
ENST00000559484.1
ENST00000354181.3 ENST00000558589.1 ENST00000458406.2 |
SLC12A6
|
solute carrier family 12 (potassium/chloride transporter), member 6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXD1_FOXO1_FOXO6_FOXG1_FOXP1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.5 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.3 | 1.1 | GO:0072308 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 0.3 | 1.3 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.3 | 1.3 | GO:0052250 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.3 | 1.5 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.2 | 0.8 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.2 | 0.5 | GO:0045081 | negative regulation of interleukin-10 biosynthetic process(GO:0045081) |
| 0.2 | 0.5 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.2 | 1.8 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.1 | 0.7 | GO:0099552 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.1 | 0.8 | GO:0015887 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) |
| 0.1 | 0.3 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.1 | 0.7 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.1 | 1.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.1 | 0.4 | GO:1903517 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) negative regulation of single strand break repair(GO:1903517) negative regulation of beta-galactosidase activity(GO:1903770) telomere single strand break repair(GO:1903823) negative regulation of telomere single strand break repair(GO:1903824) |
| 0.1 | 0.6 | GO:2000669 | negative regulation of dendritic cell apoptotic process(GO:2000669) |
| 0.1 | 0.1 | GO:0014874 | response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to stimulus involved in regulation of muscle adaptation(GO:0014874) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 | 0.3 | GO:0045575 | basophil activation involved in immune response(GO:0002276) basophil activation(GO:0045575) |
| 0.1 | 0.2 | GO:0072134 | nephrogenic mesenchyme morphogenesis(GO:0072134) |
| 0.1 | 0.4 | GO:0035574 | histone H4-K20 demethylation(GO:0035574) |
| 0.1 | 0.3 | GO:0043315 | positive regulation of neutrophil degranulation(GO:0043315) cellular response to gravity(GO:0071258) positive regulation of neutrophil activation(GO:1902565) regulation of transcytosis(GO:1904298) positive regulation of transcytosis(GO:1904300) regulation of maternal process involved in parturition(GO:1904301) positive regulation of maternal process involved in parturition(GO:1904303) response to 2-O-acetyl-1-O-hexadecyl-sn-glycero-3-phosphocholine(GO:1904316) cellular response to 2-O-acetyl-1-O-hexadecyl-sn-glycero-3-phosphocholine(GO:1904317) |
| 0.1 | 0.3 | GO:1903216 | regulation of protein processing involved in protein targeting to mitochondrion(GO:1903216) negative regulation of protein processing involved in protein targeting to mitochondrion(GO:1903217) |
| 0.1 | 0.3 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.1 | 0.2 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.1 | 0.4 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.1 | 0.3 | GO:0045209 | MAPK phosphatase export from nucleus(GO:0045208) MAPK phosphatase export from nucleus, leptomycin B sensitive(GO:0045209) |
| 0.1 | 0.4 | GO:0071477 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.1 | 0.5 | GO:1904721 | negative regulation of mRNA cleavage(GO:0031438) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.1 | 0.5 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.1 | 0.7 | GO:0006335 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.4 | GO:0030421 | defecation(GO:0030421) |
| 0.1 | 0.5 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.1 | 0.3 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.1 | 0.4 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.1 | 0.2 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.1 | 0.2 | GO:1902725 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) negative regulation of satellite cell differentiation(GO:1902725) |
| 0.1 | 0.4 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.1 | 0.3 | GO:0051029 | rRNA transport(GO:0051029) |
| 0.1 | 0.3 | GO:0018032 | protein amidation(GO:0018032) |
| 0.1 | 0.2 | GO:1904897 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.1 | 0.3 | GO:2001151 | regulation of renal water transport(GO:2001151) positive regulation of renal water transport(GO:2001153) |
| 0.0 | 0.7 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.2 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.0 | 0.3 | GO:0001712 | ectoderm formation(GO:0001705) ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.6 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.3 | GO:0009597 | detection of virus(GO:0009597) |
| 0.0 | 0.0 | GO:0051025 | negative regulation of immunoglobulin secretion(GO:0051025) |
| 0.0 | 0.6 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.4 | GO:0038129 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 0.3 | GO:0045354 | negative regulation of interferon-alpha production(GO:0032687) interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) |
| 0.0 | 0.1 | GO:0033037 | polysaccharide localization(GO:0033037) |
| 0.0 | 0.2 | GO:0010716 | negative regulation of extracellular matrix disassembly(GO:0010716) |
| 0.0 | 1.5 | GO:0035024 | negative regulation of Rho protein signal transduction(GO:0035024) |
| 0.0 | 0.6 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 0.2 | GO:0002035 | brain renin-angiotensin system(GO:0002035) angiotensin-mediated drinking behavior(GO:0003051) |
| 0.0 | 0.3 | GO:1902952 | positive regulation of dendritic spine maintenance(GO:1902952) |
| 0.0 | 0.2 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 0.0 | 0.1 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.0 | 0.2 | GO:1904499 | glial cell fate determination(GO:0007403) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of chromatin-mediated maintenance of transcription(GO:1904499) positive regulation of chromatin-mediated maintenance of transcription(GO:1904501) regulation of euchromatin binding(GO:1904793) |
| 0.0 | 0.3 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.8 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 1.1 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.1 | GO:0072695 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.0 | 0.2 | GO:0006696 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.0 | 0.6 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.0 | 0.1 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.0 | 0.3 | GO:2001205 | TNFSF11-mediated signaling pathway(GO:0071847) negative regulation of osteoclast development(GO:2001205) |
| 0.0 | 0.5 | GO:1901898 | negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.5 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.2 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.3 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.4 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.1 | GO:0042704 | detection of oxygen(GO:0003032) uterine wall breakdown(GO:0042704) |
| 0.0 | 0.2 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.2 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 | 0.4 | GO:0090232 | positive regulation of spindle checkpoint(GO:0090232) |
| 0.0 | 0.3 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.4 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 | 0.4 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.0 | 0.6 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.1 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.2 | GO:0002725 | negative regulation of T cell cytokine production(GO:0002725) regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.1 | GO:1902263 | apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.0 | 0.4 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.8 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.5 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.0 | 0.1 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.0 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.1 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.0 | 0.2 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.0 | 0.8 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.2 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.6 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 0.1 | GO:2001016 | positive regulation of skeletal muscle cell differentiation(GO:2001016) |
| 0.0 | 0.4 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.5 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 | 1.4 | GO:0003254 | regulation of membrane depolarization(GO:0003254) |
| 0.0 | 0.1 | GO:0060414 | optic placode formation involved in camera-type eye formation(GO:0046619) aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.1 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.1 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.1 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.2 | GO:0071988 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.0 | 0.1 | GO:0014028 | notochord formation(GO:0014028) |
| 0.0 | 0.1 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.1 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.0 | 0.1 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.1 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.1 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.0 | 0.2 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 | 0.1 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.0 | 0.2 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.2 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.2 | GO:0033210 | leptin-mediated signaling pathway(GO:0033210) adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.1 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.0 | 0.1 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.0 | 0.1 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.0 | 0.1 | GO:1904075 | trophectodermal cell proliferation(GO:0001834) regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.0 | 0.1 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.0 | 0.1 | GO:0070343 | white fat cell proliferation(GO:0070343) positive regulation of fat cell proliferation(GO:0070346) regulation of white fat cell proliferation(GO:0070350) |
| 0.0 | 0.7 | GO:1902751 | positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.0 | 1.1 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.1 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.1 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.1 | GO:1904016 | response to Thyroglobulin triiodothyronine(GO:1904016) |
| 0.0 | 0.0 | GO:0015766 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.0 | GO:0035553 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.0 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.0 | 0.2 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.2 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.0 | GO:0098507 | polynucleotide 5' dephosphorylation(GO:0098507) |
| 0.0 | 0.2 | GO:0051770 | positive regulation of nitric-oxide synthase biosynthetic process(GO:0051770) |
| 0.0 | 0.1 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.0 | 0.0 | GO:0021778 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.1 | GO:0030952 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) establishment or maintenance of cytoskeleton polarity(GO:0030952) |
| 0.0 | 0.3 | GO:0060065 | uterus development(GO:0060065) |
| 0.0 | 0.4 | GO:0006625 | protein targeting to peroxisome(GO:0006625) protein localization to peroxisome(GO:0072662) establishment of protein localization to peroxisome(GO:0072663) |
| 0.0 | 0.1 | GO:0097119 | postsynaptic density protein 95 clustering(GO:0097119) |
| 0.0 | 0.1 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.4 | GO:0036152 | phosphatidylethanolamine acyl-chain remodeling(GO:0036152) |
| 0.0 | 0.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.2 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.0 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.0 | 0.1 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.0 | 0.2 | GO:0046069 | cGMP catabolic process(GO:0046069) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 0.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 0.2 | GO:0044218 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.1 | 0.2 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 0.4 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 0.7 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 0.3 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.2 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.4 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 1.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.8 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.0 | 0.5 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.6 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.3 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.9 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.5 | GO:0033643 | host cell part(GO:0033643) |
| 0.0 | 0.5 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.1 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.5 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.3 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 1.1 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.1 | GO:0016938 | kinesin I complex(GO:0016938) |
| 0.0 | 0.1 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.4 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.2 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.0 | 0.6 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.3 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.2 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.6 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.1 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.0 | 0.2 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 0.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.2 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.0 | GO:0005940 | septin ring(GO:0005940) septin collar(GO:0032173) |
| 0.0 | 0.1 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.0 | 1.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.1 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.0 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.1 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.1 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.3 | 0.8 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.2 | 0.5 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.2 | 0.5 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.2 | 0.9 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 0.7 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.1 | 0.3 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.1 | 0.6 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.1 | 0.8 | GO:0008523 | sodium-dependent multivitamin transmembrane transporter activity(GO:0008523) |
| 0.1 | 1.3 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.1 | 0.4 | GO:0035575 | histone demethylase activity (H4-K20 specific)(GO:0035575) |
| 0.1 | 0.4 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.1 | 0.3 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.1 | 0.2 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.1 | 0.3 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.1 | 0.2 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.1 | 0.2 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.1 | 0.3 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.1 | 0.2 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 0.5 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.1 | 0.2 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.1 | 0.6 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.1 | 1.5 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 0.2 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.1 | 0.2 | GO:0030617 | transforming growth factor beta receptor, inhibitory cytoplasmic mediator activity(GO:0030617) |
| 0.1 | 0.3 | GO:0004598 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.0 | 0.4 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.3 | GO:0004992 | platelet activating factor receptor activity(GO:0004992) |
| 0.0 | 0.9 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.2 | GO:0051996 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.0 | 0.5 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.6 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 1.2 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.3 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.4 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.2 | GO:0008267 | poly-glutamine tract binding(GO:0008267) |
| 0.0 | 0.3 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.4 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.4 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.3 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.0 | 0.2 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.2 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.1 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.0 | 0.4 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.4 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 0.4 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.6 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.0 | 0.2 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.1 | GO:0050659 | chondroitin 4-sulfotransferase activity(GO:0047756) N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.0 | 0.1 | GO:0005199 | structural constituent of cell wall(GO:0005199) |
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.1 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.0 | 0.1 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.0 | 0.6 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.9 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.4 | GO:0044213 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.3 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.9 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.6 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 1.9 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 1.1 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 1.2 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.3 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.1 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.2 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.4 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.0 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.0 | 0.8 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.0 | 0.1 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.0 | 0.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.0 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.3 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.1 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.0 | GO:0004651 | polynucleotide 5'-phosphatase activity(GO:0004651) |
| 0.0 | 0.2 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.2 | GO:0001083 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, RNA polymerase II basal transcription factor binding(GO:0001083) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.9 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.1 | GO:0004905 | type I interferon receptor activity(GO:0004905) |
| 0.0 | 0.0 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.6 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.2 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.1 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 1.3 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.7 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.6 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 1.0 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.3 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.4 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.5 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.9 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.0 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 0.4 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.3 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.8 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.9 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.8 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.7 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.6 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.6 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.0 | 0.5 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.6 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 1.3 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.5 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.4 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.8 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.5 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.2 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.7 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |