Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
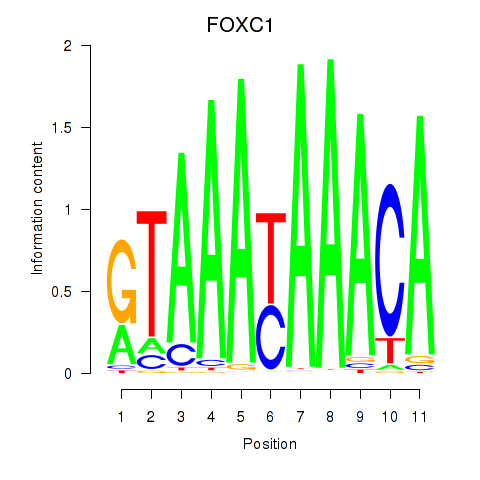
Results for FOXC1
Z-value: 0.87
Transcription factors associated with FOXC1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXC1
|
ENSG00000054598.5 | forkhead box C1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXC1 | hg19_v2_chr6_+_1610681_1610681 | -0.94 | 6.1e-02 | Click! |
Activity profile of FOXC1 motif
Sorted Z-values of FOXC1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_23261589 | 0.98 |
ENST00000524168.1
ENST00000523833.2 ENST00000519243.1 ENST00000389131.3 |
LOXL2
|
lysyl oxidase-like 2 |
| chr2_+_66918558 | 0.94 |
ENST00000435389.1
ENST00000428590.1 ENST00000412944.1 |
AC007392.3
|
AC007392.3 |
| chr22_+_46449674 | 0.85 |
ENST00000381051.2
|
FLJ27365
|
hsa-mir-4763 |
| chr6_+_84563295 | 0.79 |
ENST00000369687.1
|
RIPPLY2
|
ripply transcriptional repressor 2 |
| chr12_-_12714006 | 0.72 |
ENST00000541207.1
|
DUSP16
|
dual specificity phosphatase 16 |
| chr3_-_139195350 | 0.71 |
ENST00000232217.2
|
RBP2
|
retinol binding protein 2, cellular |
| chr8_-_123793048 | 0.61 |
ENST00000607710.1
|
RP11-44N11.2
|
RP11-44N11.2 |
| chr7_-_27219849 | 0.58 |
ENST00000396344.4
|
HOXA10
|
homeobox A10 |
| chr11_+_130184888 | 0.56 |
ENST00000602376.1
ENST00000532116.3 ENST00000602310.1 |
RP11-121M22.1
|
RP11-121M22.1 |
| chr15_+_63188009 | 0.55 |
ENST00000557900.1
|
RP11-1069G10.2
|
RP11-1069G10.2 |
| chr22_-_50970919 | 0.55 |
ENST00000329363.4
ENST00000437588.1 |
ODF3B
|
outer dense fiber of sperm tails 3B |
| chr20_+_9049303 | 0.54 |
ENST00000407043.2
ENST00000441846.1 |
PLCB4
|
phospholipase C, beta 4 |
| chr22_-_50970566 | 0.52 |
ENST00000405135.1
ENST00000401779.1 |
ODF3B
|
outer dense fiber of sperm tails 3B |
| chr11_-_104480019 | 0.52 |
ENST00000536529.1
ENST00000545630.1 ENST00000538641.1 |
RP11-886D15.1
|
RP11-886D15.1 |
| chr1_+_150122034 | 0.51 |
ENST00000025469.6
ENST00000369124.4 |
PLEKHO1
|
pleckstrin homology domain containing, family O member 1 |
| chr1_+_155099927 | 0.51 |
ENST00000368407.3
|
EFNA1
|
ephrin-A1 |
| chr12_+_43086018 | 0.51 |
ENST00000550177.1
|
RP11-25I15.3
|
RP11-25I15.3 |
| chr11_-_121986923 | 0.48 |
ENST00000560104.1
|
BLID
|
BH3-like motif containing, cell death inducer |
| chr2_+_12858355 | 0.46 |
ENST00000405331.3
|
TRIB2
|
tribbles pseudokinase 2 |
| chr12_-_12714025 | 0.45 |
ENST00000539940.1
|
DUSP16
|
dual specificity phosphatase 16 |
| chr12_+_10365082 | 0.45 |
ENST00000545859.1
|
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr22_-_50970506 | 0.44 |
ENST00000428989.2
ENST00000403326.1 |
ODF3B
|
outer dense fiber of sperm tails 3B |
| chr19_+_29493486 | 0.43 |
ENST00000589821.1
|
CTD-2081K17.2
|
CTD-2081K17.2 |
| chr17_+_7533439 | 0.39 |
ENST00000441599.2
ENST00000380450.4 ENST00000416273.3 ENST00000575903.1 ENST00000576830.1 ENST00000571153.1 ENST00000575618.1 ENST00000576152.1 |
SHBG
|
sex hormone-binding globulin |
| chr8_-_134309823 | 0.38 |
ENST00000414097.2
|
NDRG1
|
N-myc downstream regulated 1 |
| chr6_+_47666275 | 0.37 |
ENST00000327753.3
ENST00000283303.2 |
GPR115
|
G protein-coupled receptor 115 |
| chr8_-_134309335 | 0.36 |
ENST00000522890.1
ENST00000323851.7 ENST00000518176.1 ENST00000354944.5 ENST00000537882.1 ENST00000522476.1 ENST00000518066.1 ENST00000521544.1 ENST00000518480.1 ENST00000523892.1 |
NDRG1
|
N-myc downstream regulated 1 |
| chr1_+_39670360 | 0.36 |
ENST00000494012.1
|
MACF1
|
microtubule-actin crosslinking factor 1 |
| chr5_-_115872142 | 0.36 |
ENST00000510263.1
|
SEMA6A
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr6_-_31651817 | 0.35 |
ENST00000375863.3
ENST00000375860.2 |
LY6G5C
|
lymphocyte antigen 6 complex, locus G5C |
| chr5_+_169011033 | 0.33 |
ENST00000513795.1
|
SPDL1
|
spindle apparatus coiled-coil protein 1 |
| chr6_+_34204642 | 0.32 |
ENST00000347617.6
ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1
|
high mobility group AT-hook 1 |
| chr16_-_88772670 | 0.32 |
ENST00000562544.1
|
RNF166
|
ring finger protein 166 |
| chr2_-_152146385 | 0.32 |
ENST00000414946.1
ENST00000243346.5 |
NMI
|
N-myc (and STAT) interactor |
| chr8_+_31496809 | 0.31 |
ENST00000518104.1
ENST00000519301.1 |
NRG1
|
neuregulin 1 |
| chr1_+_22778337 | 0.30 |
ENST00000404138.1
ENST00000400239.2 ENST00000375647.4 ENST00000374651.4 |
ZBTB40
|
zinc finger and BTB domain containing 40 |
| chr5_+_95385819 | 0.30 |
ENST00000507997.1
|
RP11-254I22.1
|
RP11-254I22.1 |
| chr3_-_135916073 | 0.30 |
ENST00000481989.1
|
MSL2
|
male-specific lethal 2 homolog (Drosophila) |
| chr4_-_99578789 | 0.30 |
ENST00000511651.1
ENST00000505184.1 |
TSPAN5
|
tetraspanin 5 |
| chr1_+_220701456 | 0.30 |
ENST00000366918.4
ENST00000402574.1 |
MARK1
|
MAP/microtubule affinity-regulating kinase 1 |
| chr12_-_116714564 | 0.29 |
ENST00000548743.1
|
MED13L
|
mediator complex subunit 13-like |
| chr1_+_43855545 | 0.29 |
ENST00000372450.4
ENST00000310739.4 |
SZT2
|
seizure threshold 2 homolog (mouse) |
| chr3_-_178865747 | 0.29 |
ENST00000435560.1
|
RP11-360P21.2
|
RP11-360P21.2 |
| chr6_-_42418999 | 0.28 |
ENST00000340840.2
ENST00000354325.2 |
TRERF1
|
transcriptional regulating factor 1 |
| chr17_+_79953310 | 0.28 |
ENST00000582355.2
|
ASPSCR1
|
alveolar soft part sarcoma chromosome region, candidate 1 |
| chr5_-_24645078 | 0.28 |
ENST00000264463.4
|
CDH10
|
cadherin 10, type 2 (T2-cadherin) |
| chr4_+_86396321 | 0.28 |
ENST00000503995.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr6_+_15401075 | 0.27 |
ENST00000541660.1
|
JARID2
|
jumonji, AT rich interactive domain 2 |
| chr11_+_46639071 | 0.27 |
ENST00000580238.1
ENST00000581416.1 ENST00000529655.1 ENST00000533325.1 ENST00000581438.1 ENST00000583249.1 ENST00000530500.1 ENST00000526508.1 ENST00000578626.1 ENST00000577256.1 ENST00000524625.1 ENST00000582547.1 ENST00000359513.4 ENST00000528494.1 |
ATG13
|
autophagy related 13 |
| chr9_+_134103496 | 0.27 |
ENST00000498010.1
ENST00000476004.1 ENST00000528406.1 |
NUP214
|
nucleoporin 214kDa |
| chr12_+_59989918 | 0.27 |
ENST00000547379.1
ENST00000549465.1 |
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr11_-_66112555 | 0.26 |
ENST00000425825.2
ENST00000359957.3 |
BRMS1
|
breast cancer metastasis suppressor 1 |
| chr3_-_49131614 | 0.26 |
ENST00000450685.1
|
QRICH1
|
glutamine-rich 1 |
| chr11_+_8040739 | 0.25 |
ENST00000534099.1
|
TUB
|
tubby bipartite transcription factor |
| chr2_-_224467002 | 0.25 |
ENST00000421386.1
ENST00000433889.1 |
SCG2
|
secretogranin II |
| chr12_+_54378849 | 0.25 |
ENST00000515593.1
|
HOXC10
|
homeobox C10 |
| chr17_+_72426891 | 0.25 |
ENST00000392627.1
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr7_+_120590803 | 0.25 |
ENST00000315870.5
ENST00000339121.5 ENST00000445699.1 |
ING3
|
inhibitor of growth family, member 3 |
| chr15_+_74509530 | 0.24 |
ENST00000321288.5
|
CCDC33
|
coiled-coil domain containing 33 |
| chr11_+_46638805 | 0.24 |
ENST00000434074.1
ENST00000312040.4 ENST00000451945.1 |
ATG13
|
autophagy related 13 |
| chr2_+_58655461 | 0.24 |
ENST00000429095.1
ENST00000429664.1 ENST00000452840.1 |
AC007092.1
|
long intergenic non-protein coding RNA 1122 |
| chr3_-_71632894 | 0.24 |
ENST00000493089.1
|
FOXP1
|
forkhead box P1 |
| chr7_-_99698338 | 0.23 |
ENST00000354230.3
ENST00000425308.1 |
MCM7
|
minichromosome maintenance complex component 7 |
| chr6_+_30130969 | 0.23 |
ENST00000376694.4
|
TRIM15
|
tripartite motif containing 15 |
| chr15_+_67418047 | 0.23 |
ENST00000540846.2
|
SMAD3
|
SMAD family member 3 |
| chr9_-_139258159 | 0.23 |
ENST00000371739.3
|
DNLZ
|
DNL-type zinc finger |
| chr15_+_80351910 | 0.23 |
ENST00000261749.6
ENST00000561060.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr10_+_71561649 | 0.23 |
ENST00000398978.3
ENST00000354547.3 ENST00000357811.3 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr2_+_173724771 | 0.22 |
ENST00000538974.1
ENST00000540783.1 |
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr10_-_97200772 | 0.22 |
ENST00000371241.1
ENST00000354106.3 ENST00000371239.1 ENST00000361941.3 ENST00000277982.5 ENST00000371245.3 |
SORBS1
|
sorbin and SH3 domain containing 1 |
| chr4_+_68424434 | 0.22 |
ENST00000265404.2
ENST00000396225.1 |
STAP1
|
signal transducing adaptor family member 1 |
| chr3_-_129279894 | 0.22 |
ENST00000506979.1
|
PLXND1
|
plexin D1 |
| chr9_-_139258235 | 0.22 |
ENST00000371738.3
|
DNLZ
|
DNL-type zinc finger |
| chr12_+_93096619 | 0.21 |
ENST00000397833.3
|
C12orf74
|
chromosome 12 open reading frame 74 |
| chr1_-_153931052 | 0.21 |
ENST00000368630.3
ENST00000368633.1 |
CRTC2
|
CREB regulated transcription coactivator 2 |
| chr9_-_20382446 | 0.21 |
ENST00000380321.1
|
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr3_-_49131788 | 0.21 |
ENST00000395443.2
ENST00000411682.1 |
QRICH1
|
glutamine-rich 1 |
| chr12_+_93096759 | 0.21 |
ENST00000544406.2
|
C12orf74
|
chromosome 12 open reading frame 74 |
| chr2_+_210518057 | 0.21 |
ENST00000452717.1
|
MAP2
|
microtubule-associated protein 2 |
| chr8_-_145754428 | 0.20 |
ENST00000527462.1
ENST00000313465.5 ENST00000524821.1 |
C8orf82
|
chromosome 8 open reading frame 82 |
| chr10_+_69869237 | 0.19 |
ENST00000373675.3
|
MYPN
|
myopalladin |
| chr1_-_57045228 | 0.19 |
ENST00000371250.3
|
PPAP2B
|
phosphatidic acid phosphatase type 2B |
| chr2_-_25451065 | 0.19 |
ENST00000606328.1
|
RP11-458N5.1
|
RP11-458N5.1 |
| chr10_+_71561630 | 0.19 |
ENST00000398974.3
ENST00000398971.3 ENST00000398968.3 ENST00000398966.3 ENST00000398964.3 ENST00000398969.3 ENST00000356340.3 ENST00000398972.3 ENST00000398973.3 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr5_+_40841276 | 0.19 |
ENST00000254691.5
|
CARD6
|
caspase recruitment domain family, member 6 |
| chr11_-_65359947 | 0.18 |
ENST00000597463.1
|
AP001362.1
|
Uncharacterized protein |
| chr11_+_10476851 | 0.18 |
ENST00000396553.2
|
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr6_+_53794780 | 0.18 |
ENST00000505762.1
ENST00000511369.1 ENST00000431554.2 |
MLIP
RP11-411K7.1
|
muscular LMNA-interacting protein RP11-411K7.1 |
| chr11_-_46638720 | 0.18 |
ENST00000326737.3
|
HARBI1
|
harbinger transposase derived 1 |
| chr15_+_74911430 | 0.18 |
ENST00000562670.1
ENST00000564096.1 |
CLK3
|
CDC-like kinase 3 |
| chr8_+_11666649 | 0.17 |
ENST00000528643.1
ENST00000525777.1 |
FDFT1
|
farnesyl-diphosphate farnesyltransferase 1 |
| chr5_-_36241900 | 0.17 |
ENST00000381937.4
ENST00000514504.1 |
NADK2
|
NAD kinase 2, mitochondrial |
| chr2_-_207023918 | 0.17 |
ENST00000455934.2
ENST00000449699.1 ENST00000454195.1 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr1_-_94079648 | 0.17 |
ENST00000370247.3
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr13_-_36429763 | 0.17 |
ENST00000379893.1
|
DCLK1
|
doublecortin-like kinase 1 |
| chr5_+_150040403 | 0.17 |
ENST00000517768.1
ENST00000297130.4 |
MYOZ3
|
myozenin 3 |
| chr2_+_210517895 | 0.17 |
ENST00000447185.1
|
MAP2
|
microtubule-associated protein 2 |
| chr5_+_112312416 | 0.17 |
ENST00000389063.2
|
DCP2
|
decapping mRNA 2 |
| chr10_-_4285923 | 0.17 |
ENST00000418372.1
ENST00000608792.1 |
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr9_+_71944241 | 0.17 |
ENST00000257515.8
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr7_-_14028488 | 0.17 |
ENST00000405358.4
|
ETV1
|
ets variant 1 |
| chr1_-_178840157 | 0.17 |
ENST00000367629.1
ENST00000234816.2 |
ANGPTL1
|
angiopoietin-like 1 |
| chr19_-_4831701 | 0.17 |
ENST00000248244.5
|
TICAM1
|
toll-like receptor adaptor molecule 1 |
| chr13_+_27825446 | 0.16 |
ENST00000311549.6
|
RPL21
|
ribosomal protein L21 |
| chr7_-_27219632 | 0.16 |
ENST00000470747.4
|
RP1-170O19.20
|
Uncharacterized protein |
| chr4_+_166794383 | 0.16 |
ENST00000061240.2
ENST00000507499.1 |
TLL1
|
tolloid-like 1 |
| chr19_+_29493456 | 0.16 |
ENST00000591143.1
ENST00000592653.1 |
CTD-2081K17.2
|
CTD-2081K17.2 |
| chr8_-_116673894 | 0.16 |
ENST00000395713.2
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr18_+_19749386 | 0.16 |
ENST00000269216.3
|
GATA6
|
GATA binding protein 6 |
| chr4_+_86396265 | 0.16 |
ENST00000395184.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr10_+_52751010 | 0.16 |
ENST00000373985.1
|
PRKG1
|
protein kinase, cGMP-dependent, type I |
| chr12_+_122516626 | 0.16 |
ENST00000319080.7
|
MLXIP
|
MLX interacting protein |
| chr16_-_4817129 | 0.16 |
ENST00000545009.1
ENST00000219478.6 |
ZNF500
|
zinc finger protein 500 |
| chr13_-_36050819 | 0.15 |
ENST00000379919.4
|
MAB21L1
|
mab-21-like 1 (C. elegans) |
| chrX_-_19689106 | 0.15 |
ENST00000379716.1
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr2_-_208030295 | 0.15 |
ENST00000458272.1
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr1_+_199996733 | 0.15 |
ENST00000236914.3
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr7_+_123488124 | 0.15 |
ENST00000476325.1
|
HYAL4
|
hyaluronoglucosaminidase 4 |
| chr1_-_43855444 | 0.14 |
ENST00000372455.4
|
MED8
|
mediator complex subunit 8 |
| chr10_-_31288398 | 0.14 |
ENST00000538351.2
|
ZNF438
|
zinc finger protein 438 |
| chr10_+_71562180 | 0.14 |
ENST00000517713.1
ENST00000522165.1 ENST00000520133.1 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr9_-_4299874 | 0.14 |
ENST00000381971.3
ENST00000477901.1 |
GLIS3
|
GLIS family zinc finger 3 |
| chr14_-_24020858 | 0.14 |
ENST00000419474.3
|
ZFHX2
|
zinc finger homeobox 2 |
| chr10_+_114710425 | 0.14 |
ENST00000352065.5
ENST00000369395.1 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr12_+_59989791 | 0.13 |
ENST00000552432.1
|
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr14_+_37126765 | 0.13 |
ENST00000402703.2
|
PAX9
|
paired box 9 |
| chr16_-_740354 | 0.13 |
ENST00000293883.4
|
WDR24
|
WD repeat domain 24 |
| chr19_-_51141196 | 0.13 |
ENST00000338916.4
|
SYT3
|
synaptotagmin III |
| chr16_+_30064411 | 0.13 |
ENST00000338110.5
|
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr1_+_39670423 | 0.13 |
ENST00000536367.1
|
MACF1
|
microtubule-actin crosslinking factor 1 |
| chr6_-_134638767 | 0.13 |
ENST00000524929.1
|
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr2_-_207024134 | 0.12 |
ENST00000457011.1
ENST00000440274.1 ENST00000432169.1 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr5_-_58571935 | 0.12 |
ENST00000503258.1
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr2_+_85981008 | 0.12 |
ENST00000306279.3
|
ATOH8
|
atonal homolog 8 (Drosophila) |
| chr7_-_139876812 | 0.12 |
ENST00000397560.2
|
JHDM1D
|
lysine (K)-specific demethylase 7A |
| chr14_+_23340822 | 0.12 |
ENST00000359591.4
|
LRP10
|
low density lipoprotein receptor-related protein 10 |
| chr17_+_28268623 | 0.12 |
ENST00000394835.3
ENST00000320856.5 ENST00000394832.2 ENST00000378738.3 |
EFCAB5
|
EF-hand calcium binding domain 5 |
| chr16_+_30064444 | 0.12 |
ENST00000395248.1
ENST00000566897.1 ENST00000568435.1 |
ALDOA
|
aldolase A, fructose-bisphosphate |
| chr1_+_18958008 | 0.12 |
ENST00000420770.2
ENST00000400661.3 |
PAX7
|
paired box 7 |
| chr6_-_5260963 | 0.12 |
ENST00000464010.1
ENST00000468929.1 ENST00000480566.1 |
LYRM4
|
LYR motif containing 4 |
| chr21_+_38071430 | 0.12 |
ENST00000290399.6
|
SIM2
|
single-minded family bHLH transcription factor 2 |
| chr7_+_80231466 | 0.12 |
ENST00000309881.7
ENST00000534394.1 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr4_+_185395947 | 0.12 |
ENST00000605834.1
|
RP11-326I11.3
|
RP11-326I11.3 |
| chr1_+_197886461 | 0.12 |
ENST00000367388.3
ENST00000337020.2 ENST00000367387.4 |
LHX9
|
LIM homeobox 9 |
| chr7_+_136553370 | 0.12 |
ENST00000445907.2
|
CHRM2
|
cholinergic receptor, muscarinic 2 |
| chr2_-_208030647 | 0.12 |
ENST00000309446.6
|
KLF7
|
Kruppel-like factor 7 (ubiquitous) |
| chr3_+_57875711 | 0.11 |
ENST00000442599.2
|
SLMAP
|
sarcolemma associated protein |
| chr18_+_32073253 | 0.11 |
ENST00000283365.9
ENST00000596745.1 ENST00000315456.6 |
DTNA
|
dystrobrevin, alpha |
| chr9_-_89562104 | 0.11 |
ENST00000298743.7
|
GAS1
|
growth arrest-specific 1 |
| chr11_-_108464465 | 0.11 |
ENST00000525344.1
|
EXPH5
|
exophilin 5 |
| chr6_+_32121908 | 0.11 |
ENST00000375143.2
ENST00000424499.1 |
PPT2
|
palmitoyl-protein thioesterase 2 |
| chrX_-_19688475 | 0.11 |
ENST00000541422.1
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr9_+_127054254 | 0.11 |
ENST00000422297.1
|
NEK6
|
NIMA-related kinase 6 |
| chr6_-_56716686 | 0.11 |
ENST00000520645.1
|
DST
|
dystonin |
| chr1_-_94312706 | 0.11 |
ENST00000370244.1
|
BCAR3
|
breast cancer anti-estrogen resistance 3 |
| chr17_-_48546232 | 0.11 |
ENST00000258969.4
|
CHAD
|
chondroadherin |
| chr9_-_98268883 | 0.11 |
ENST00000551630.1
ENST00000548420.1 |
PTCH1
|
patched 1 |
| chr4_-_74486347 | 0.11 |
ENST00000342081.3
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr5_-_36242119 | 0.11 |
ENST00000511088.1
ENST00000282512.3 ENST00000506945.1 |
NADK2
|
NAD kinase 2, mitochondrial |
| chr12_-_31479107 | 0.11 |
ENST00000542983.1
|
FAM60A
|
family with sequence similarity 60, member A |
| chr17_+_40925454 | 0.11 |
ENST00000253794.2
ENST00000590339.1 ENST00000589520.1 |
VPS25
|
vacuolar protein sorting 25 homolog (S. cerevisiae) |
| chr11_+_10477733 | 0.10 |
ENST00000528723.1
|
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr1_+_226411319 | 0.10 |
ENST00000542034.1
ENST00000366810.5 |
MIXL1
|
Mix paired-like homeobox |
| chr2_+_207024306 | 0.10 |
ENST00000236957.5
ENST00000392221.1 ENST00000392222.2 ENST00000445505.1 |
EEF1B2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr1_+_25664408 | 0.10 |
ENST00000374358.4
|
TMEM50A
|
transmembrane protein 50A |
| chr15_+_80351977 | 0.10 |
ENST00000559157.1
ENST00000561012.1 ENST00000564367.1 ENST00000558494.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr4_+_57276661 | 0.10 |
ENST00000598320.1
|
AC068620.1
|
Uncharacterized protein |
| chr1_-_159684371 | 0.10 |
ENST00000255030.5
ENST00000437342.1 ENST00000368112.1 ENST00000368111.1 ENST00000368110.1 ENST00000343919.2 |
CRP
|
C-reactive protein, pentraxin-related |
| chr4_-_185395191 | 0.10 |
ENST00000510814.1
ENST00000507523.1 ENST00000506230.1 |
IRF2
|
interferon regulatory factor 2 |
| chr3_+_72201910 | 0.10 |
ENST00000469178.1
ENST00000485404.1 |
LINC00870
|
long intergenic non-protein coding RNA 870 |
| chr12_+_10365404 | 0.10 |
ENST00000266458.5
ENST00000421801.2 ENST00000544284.1 ENST00000545047.1 ENST00000543602.1 ENST00000545887.1 |
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr11_-_108464321 | 0.10 |
ENST00000265843.4
|
EXPH5
|
exophilin 5 |
| chr16_+_72459838 | 0.10 |
ENST00000564508.1
|
AC004158.3
|
AC004158.3 |
| chr5_-_55529115 | 0.10 |
ENST00000513241.2
ENST00000341048.4 |
ANKRD55
|
ankyrin repeat domain 55 |
| chr1_+_12524965 | 0.10 |
ENST00000471923.1
|
VPS13D
|
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
| chr7_+_151038850 | 0.10 |
ENST00000355851.4
ENST00000566856.1 ENST00000470229.1 |
NUB1
|
negative regulator of ubiquitin-like proteins 1 |
| chr1_+_43855560 | 0.10 |
ENST00000562955.1
|
SZT2
|
seizure threshold 2 homolog (mouse) |
| chr2_+_173792893 | 0.10 |
ENST00000535187.1
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr21_+_39628780 | 0.10 |
ENST00000417042.1
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr21_+_39628852 | 0.10 |
ENST00000398938.2
|
KCNJ15
|
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr18_-_53303123 | 0.10 |
ENST00000569357.1
ENST00000565124.1 ENST00000398339.1 |
TCF4
|
transcription factor 4 |
| chr17_-_48546324 | 0.09 |
ENST00000508540.1
|
CHAD
|
chondroadherin |
| chr14_+_69726864 | 0.09 |
ENST00000448469.3
|
GALNT16
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 16 |
| chr17_+_9745786 | 0.09 |
ENST00000304773.5
|
GLP2R
|
glucagon-like peptide 2 receptor |
| chr14_+_69726656 | 0.09 |
ENST00000337827.4
|
GALNT16
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 16 |
| chr3_-_134092561 | 0.09 |
ENST00000510560.1
ENST00000504234.1 ENST00000515172.1 |
AMOTL2
|
angiomotin like 2 |
| chr5_+_159614374 | 0.09 |
ENST00000393980.4
|
FABP6
|
fatty acid binding protein 6, ileal |
| chr3_-_16524357 | 0.09 |
ENST00000432519.1
|
RFTN1
|
raftlin, lipid raft linker 1 |
| chr14_-_20774092 | 0.09 |
ENST00000423949.2
ENST00000553828.1 ENST00000258821.3 |
TTC5
|
tetratricopeptide repeat domain 5 |
| chr13_-_99667960 | 0.09 |
ENST00000448493.2
|
DOCK9
|
dedicator of cytokinesis 9 |
| chr6_+_32121789 | 0.09 |
ENST00000437001.2
ENST00000375137.2 |
PPT2
|
palmitoyl-protein thioesterase 2 |
| chr1_+_56046710 | 0.09 |
ENST00000422374.1
|
RP11-466L17.1
|
RP11-466L17.1 |
| chr9_+_127054217 | 0.08 |
ENST00000394199.2
ENST00000546191.1 |
NEK6
|
NIMA-related kinase 6 |
| chr16_-_740419 | 0.08 |
ENST00000248142.6
|
WDR24
|
WD repeat domain 24 |
| chr7_+_28452130 | 0.08 |
ENST00000357727.2
|
CREB5
|
cAMP responsive element binding protein 5 |
| chr2_+_105050794 | 0.08 |
ENST00000429464.1
ENST00000414442.1 ENST00000447380.1 |
AC013402.2
|
long intergenic non-protein coding RNA 1102 |
| chr1_+_204839959 | 0.08 |
ENST00000404076.1
|
NFASC
|
neurofascin |
| chr4_-_83483395 | 0.08 |
ENST00000515780.2
|
TMEM150C
|
transmembrane protein 150C |
| chr4_+_71588372 | 0.08 |
ENST00000536664.1
|
RUFY3
|
RUN and FYVE domain containing 3 |
| chr22_+_41487711 | 0.08 |
ENST00000263253.7
|
EP300
|
E1A binding protein p300 |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXC1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0045208 | MAPK phosphatase export from nucleus(GO:0045208) MAPK phosphatase export from nucleus, leptomycin B sensitive(GO:0045209) |
| 0.2 | 1.0 | GO:0018277 | protein deamination(GO:0018277) |
| 0.2 | 0.5 | GO:0045081 | negative regulation of interleukin-10 biosynthetic process(GO:0045081) |
| 0.1 | 0.5 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.1 | 0.2 | GO:1903980 | negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) positive regulation of microglial cell activation(GO:1903980) |
| 0.1 | 0.8 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.1 | 0.3 | GO:0090402 | senescence-associated heterochromatin focus assembly(GO:0035986) oncogene-induced cell senescence(GO:0090402) |
| 0.1 | 0.7 | GO:0090232 | positive regulation of spindle checkpoint(GO:0090232) |
| 0.1 | 0.1 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.1 | 0.7 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.1 | 0.4 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.1 | 0.2 | GO:0007493 | endodermal cell fate determination(GO:0007493) |
| 0.1 | 0.3 | GO:0045354 | negative regulation of interferon-alpha production(GO:0032687) interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) |
| 0.1 | 0.2 | GO:1900155 | regulation of bone trabecula formation(GO:1900154) negative regulation of bone trabecula formation(GO:1900155) |
| 0.1 | 0.4 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.2 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.0 | 0.4 | GO:0038129 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 0.3 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.0 | 0.2 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.3 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.2 | GO:0008204 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.0 | 0.2 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.2 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.5 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.2 | GO:0045359 | interferon-beta biosynthetic process(GO:0045350) regulation of interferon-beta biosynthetic process(GO:0045357) positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.0 | 0.2 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) |
| 0.0 | 0.5 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.0 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.3 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) |
| 0.0 | 0.3 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.1 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.1 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.1 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.2 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.3 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.6 | GO:0098780 | response to mitochondrial depolarisation(GO:0098780) |
| 0.0 | 0.2 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.0 | 0.6 | GO:0060065 | uterus development(GO:0060065) |
| 0.0 | 0.3 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.2 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.4 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.1 | GO:2000334 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 0.0 | GO:0036518 | chemorepulsion of dopaminergic neuron axon(GO:0036518) |
| 0.0 | 0.2 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.2 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.3 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.2 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.1 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.0 | 0.1 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.1 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 0.3 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.2 | GO:0007379 | segment specification(GO:0007379) |
| 0.0 | 0.1 | GO:0021764 | amygdala development(GO:0021764) |
| 0.0 | 0.1 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 0.0 | GO:0015855 | pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.0 | 0.1 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.2 | 0.6 | GO:0030936 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.1 | 0.3 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.1 | 0.4 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.1 | 0.4 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.1 | 0.4 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 0.3 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.0 | 0.2 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.5 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.1 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.3 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.1 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.1 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.2 | GO:0061700 | Seh1-associated complex(GO:0035859) GATOR2 complex(GO:0061700) |
| 0.0 | 0.6 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 0.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.6 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.7 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.4 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.1 | 1.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.2 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.2 | GO:0051996 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.0 | 0.4 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.7 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.2 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.3 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.3 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.2 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.4 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.5 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.2 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.1 | GO:0033265 | choline binding(GO:0033265) |
| 0.0 | 0.2 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.3 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.0 | 0.6 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.7 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.3 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.1 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.2 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.4 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.3 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.5 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.1 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.0 | GO:0005350 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) glycerol channel activity(GO:0015254) urea channel activity(GO:0015265) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.0 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.0 | 0.3 | GO:0008301 | DNA binding, bending(GO:0008301) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 1.3 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.5 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.5 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.7 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |