Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
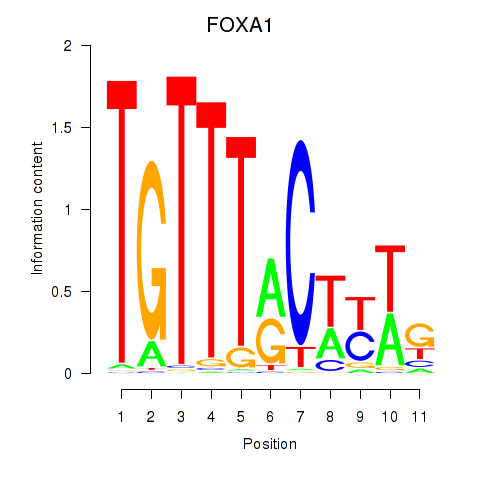
Results for FOXA1
Z-value: 1.08
Transcription factors associated with FOXA1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOXA1
|
ENSG00000129514.4 | forkhead box A1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOXA1 | hg19_v2_chr14_-_38064198_38064239 | 0.77 | 2.3e-01 | Click! |
Activity profile of FOXA1 motif
Sorted Z-values of FOXA1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_46035187 | 2.13 |
ENST00000300557.2
|
PRR15L
|
proline rich 15-like |
| chr3_-_148939598 | 1.67 |
ENST00000455472.3
|
CP
|
ceruloplasmin (ferroxidase) |
| chr9_-_4859260 | 1.30 |
ENST00000599351.1
|
AL158147.2
|
HCG2011465; Uncharacterized protein |
| chr20_-_52687059 | 1.13 |
ENST00000371435.2
ENST00000395961.3 |
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr14_-_94789663 | 1.12 |
ENST00000557225.1
ENST00000341584.3 |
SERPINA6
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 6 |
| chr10_-_90751038 | 1.07 |
ENST00000458159.1
ENST00000415557.1 ENST00000458208.1 |
ACTA2
|
actin, alpha 2, smooth muscle, aorta |
| chr3_+_186330712 | 1.04 |
ENST00000411641.2
ENST00000273784.5 |
AHSG
|
alpha-2-HS-glycoprotein |
| chr4_+_165675197 | 1.02 |
ENST00000515485.1
|
RP11-294O2.2
|
RP11-294O2.2 |
| chr21_+_17791648 | 0.95 |
ENST00000602892.1
ENST00000418813.2 ENST00000435697.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr1_-_115301235 | 0.89 |
ENST00000525878.1
|
CSDE1
|
cold shock domain containing E1, RNA-binding |
| chr15_+_96869165 | 0.84 |
ENST00000421109.2
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr14_+_38065052 | 0.84 |
ENST00000556845.1
|
TTC6
|
tetratricopeptide repeat domain 6 |
| chr2_+_162101247 | 0.75 |
ENST00000439050.1
ENST00000436506.1 |
AC009299.3
|
AC009299.3 |
| chr8_-_131028782 | 0.74 |
ENST00000519020.1
|
FAM49B
|
family with sequence similarity 49, member B |
| chr4_-_155533787 | 0.69 |
ENST00000407946.1
ENST00000405164.1 ENST00000336098.3 ENST00000393846.2 ENST00000404648.3 ENST00000443553.1 |
FGG
|
fibrinogen gamma chain |
| chr12_-_71551868 | 0.69 |
ENST00000247829.3
|
TSPAN8
|
tetraspanin 8 |
| chr12_+_96588368 | 0.68 |
ENST00000547860.1
|
ELK3
|
ELK3, ETS-domain protein (SRF accessory protein 2) |
| chr12_+_121416437 | 0.63 |
ENST00000402929.1
ENST00000535955.1 ENST00000538626.1 ENST00000543427.1 |
HNF1A
|
HNF1 homeobox A |
| chr13_-_67802549 | 0.61 |
ENST00000328454.5
ENST00000377865.2 |
PCDH9
|
protocadherin 9 |
| chr1_-_1356719 | 0.59 |
ENST00000520296.1
|
ANKRD65
|
ankyrin repeat domain 65 |
| chr17_-_57229155 | 0.58 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr1_-_1356628 | 0.56 |
ENST00000442470.1
ENST00000537107.1 |
ANKRD65
|
ankyrin repeat domain 65 |
| chr3_-_25706368 | 0.55 |
ENST00000424225.1
|
TOP2B
|
topoisomerase (DNA) II beta 180kDa |
| chr1_-_207095324 | 0.55 |
ENST00000530505.1
ENST00000367091.3 ENST00000442471.2 |
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr7_+_106415457 | 0.53 |
ENST00000490162.2
ENST00000470135.1 |
RP5-884M6.1
|
RP5-884M6.1 |
| chr12_+_10658489 | 0.52 |
ENST00000538173.1
|
EIF2S3L
|
Putative eukaryotic translation initiation factor 2 subunit 3-like protein |
| chr14_+_64971292 | 0.52 |
ENST00000358738.3
ENST00000394712.2 |
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr2_-_182545603 | 0.49 |
ENST00000295108.3
|
NEUROD1
|
neuronal differentiation 1 |
| chr18_+_72922710 | 0.47 |
ENST00000322038.5
|
TSHZ1
|
teashirt zinc finger homeobox 1 |
| chr5_-_41510725 | 0.47 |
ENST00000328457.3
|
PLCXD3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr2_+_196521845 | 0.47 |
ENST00000359634.5
ENST00000412905.1 |
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chrX_-_15402498 | 0.47 |
ENST00000297904.3
|
FIGF
|
c-fos induced growth factor (vascular endothelial growth factor D) |
| chr21_+_17791838 | 0.46 |
ENST00000453910.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr1_+_87797351 | 0.45 |
ENST00000370542.1
|
LMO4
|
LIM domain only 4 |
| chr3_-_148939835 | 0.45 |
ENST00000264613.6
|
CP
|
ceruloplasmin (ferroxidase) |
| chr1_+_198126093 | 0.45 |
ENST00000367385.4
ENST00000442588.1 ENST00000538004.1 |
NEK7
|
NIMA-related kinase 7 |
| chr7_-_84121858 | 0.44 |
ENST00000448879.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr12_-_71551652 | 0.44 |
ENST00000546561.1
|
TSPAN8
|
tetraspanin 8 |
| chr3_+_142315225 | 0.44 |
ENST00000457734.2
ENST00000483373.1 ENST00000475296.1 ENST00000495744.1 ENST00000476044.1 ENST00000461644.1 |
PLS1
|
plastin 1 |
| chr4_+_55095428 | 0.43 |
ENST00000508170.1
ENST00000512143.1 |
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr3_-_45957088 | 0.43 |
ENST00000539217.1
|
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr3_-_120068143 | 0.43 |
ENST00000295628.3
|
LRRC58
|
leucine rich repeat containing 58 |
| chr2_+_196522032 | 0.43 |
ENST00000418005.1
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr1_-_177939041 | 0.41 |
ENST00000308284.6
|
SEC16B
|
SEC16 homolog B (S. cerevisiae) |
| chr12_+_27849378 | 0.41 |
ENST00000310791.2
|
REP15
|
RAB15 effector protein |
| chr20_-_52687030 | 0.41 |
ENST00000411563.1
|
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr2_+_196521903 | 0.40 |
ENST00000541054.1
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr13_-_86373536 | 0.39 |
ENST00000400286.2
|
SLITRK6
|
SLIT and NTRK-like family, member 6 |
| chr21_-_38445011 | 0.37 |
ENST00000464265.1
ENST00000399102.1 |
PIGP
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr16_+_21608525 | 0.37 |
ENST00000567404.1
|
METTL9
|
methyltransferase like 9 |
| chrX_-_63425561 | 0.37 |
ENST00000374869.3
ENST00000330258.3 |
AMER1
|
APC membrane recruitment protein 1 |
| chr5_+_40841410 | 0.37 |
ENST00000381677.3
|
CARD6
|
caspase recruitment domain family, member 6 |
| chr4_+_56719782 | 0.36 |
ENST00000381295.2
ENST00000346134.7 ENST00000349598.6 |
EXOC1
|
exocyst complex component 1 |
| chr18_-_52626622 | 0.36 |
ENST00000591504.1
|
CCDC68
|
coiled-coil domain containing 68 |
| chr14_-_38064198 | 0.36 |
ENST00000250448.2
|
FOXA1
|
forkhead box A1 |
| chr19_-_15443318 | 0.36 |
ENST00000360016.5
|
BRD4
|
bromodomain containing 4 |
| chr7_-_105332084 | 0.35 |
ENST00000472195.1
|
ATXN7L1
|
ataxin 7-like 1 |
| chr3_+_160394940 | 0.35 |
ENST00000320767.2
|
ARL14
|
ADP-ribosylation factor-like 14 |
| chr15_+_69854027 | 0.34 |
ENST00000498938.2
|
RP11-279F6.1
|
RP11-279F6.1 |
| chr3_+_189349162 | 0.34 |
ENST00000264731.3
ENST00000382063.4 ENST00000418709.2 ENST00000320472.5 ENST00000392460.3 ENST00000440651.2 |
TP63
|
tumor protein p63 |
| chr2_-_175202151 | 0.33 |
ENST00000595354.1
|
AC018470.1
|
Uncharacterized protein FLJ46347 |
| chr1_+_101361782 | 0.32 |
ENST00000357650.4
|
SLC30A7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr5_+_95998714 | 0.32 |
ENST00000506811.1
ENST00000514055.1 |
CAST
|
calpastatin |
| chr8_-_131028869 | 0.32 |
ENST00000518283.1
ENST00000519110.1 |
FAM49B
|
family with sequence similarity 49, member B |
| chr12_-_120765565 | 0.32 |
ENST00000423423.3
ENST00000308366.4 |
PLA2G1B
|
phospholipase A2, group IB (pancreas) |
| chr3_+_141106643 | 0.31 |
ENST00000514251.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr2_+_172950227 | 0.31 |
ENST00000341900.6
|
DLX1
|
distal-less homeobox 1 |
| chr2_-_3521518 | 0.31 |
ENST00000382093.5
|
ADI1
|
acireductone dioxygenase 1 |
| chr5_+_156712372 | 0.31 |
ENST00000541131.1
|
CYFIP2
|
cytoplasmic FMR1 interacting protein 2 |
| chr10_-_32217717 | 0.30 |
ENST00000396144.4
ENST00000375245.4 ENST00000344936.2 ENST00000375250.5 |
ARHGAP12
|
Rho GTPase activating protein 12 |
| chr18_+_56530136 | 0.29 |
ENST00000591083.1
|
ZNF532
|
zinc finger protein 532 |
| chr5_-_41510656 | 0.29 |
ENST00000377801.3
|
PLCXD3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr6_+_108977520 | 0.28 |
ENST00000540898.1
|
FOXO3
|
forkhead box O3 |
| chr3_-_45957534 | 0.27 |
ENST00000536047.1
|
LZTFL1
|
leucine zipper transcription factor-like 1 |
| chr15_+_38964048 | 0.27 |
ENST00000560203.1
ENST00000557946.1 |
RP11-275I4.2
|
RP11-275I4.2 |
| chr5_+_95998673 | 0.27 |
ENST00000514845.1
|
CAST
|
calpastatin |
| chr20_+_33563206 | 0.26 |
ENST00000262873.7
|
MYH7B
|
myosin, heavy chain 7B, cardiac muscle, beta |
| chr4_-_40516560 | 0.26 |
ENST00000513473.1
|
RBM47
|
RNA binding motif protein 47 |
| chr3_-_194030493 | 0.25 |
ENST00000456816.1
ENST00000414120.1 ENST00000429578.1 |
LINC00887
|
long intergenic non-protein coding RNA 887 |
| chr2_+_47630255 | 0.25 |
ENST00000406134.1
|
MSH2
|
mutS homolog 2 |
| chr12_+_96588279 | 0.25 |
ENST00000552142.1
|
ELK3
|
ELK3, ETS-domain protein (SRF accessory protein 2) |
| chr12_-_78934441 | 0.25 |
ENST00000546865.1
ENST00000547089.1 |
RP11-171L9.1
|
RP11-171L9.1 |
| chrX_+_129473859 | 0.24 |
ENST00000424447.1
|
SLC25A14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr12_-_106697974 | 0.24 |
ENST00000553039.1
|
CKAP4
|
cytoskeleton-associated protein 4 |
| chr14_+_64970662 | 0.24 |
ENST00000556965.1
ENST00000554015.1 |
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr6_+_10528560 | 0.23 |
ENST00000379597.3
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr17_+_8924837 | 0.23 |
ENST00000173229.2
|
NTN1
|
netrin 1 |
| chr4_-_164534657 | 0.23 |
ENST00000339875.5
|
MARCH1
|
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr6_+_74405501 | 0.22 |
ENST00000437994.2
ENST00000422508.2 |
CD109
|
CD109 molecule |
| chr3_+_108321623 | 0.22 |
ENST00000497905.1
ENST00000463306.1 |
DZIP3
|
DAZ interacting zinc finger protein 3 |
| chr1_+_246729815 | 0.22 |
ENST00000366511.1
|
CNST
|
consortin, connexin sorting protein |
| chr13_+_78109884 | 0.22 |
ENST00000377246.3
ENST00000349847.3 |
SCEL
|
sciellin |
| chr4_-_174256276 | 0.22 |
ENST00000296503.5
|
HMGB2
|
high mobility group box 2 |
| chr6_+_74405804 | 0.22 |
ENST00000287097.5
|
CD109
|
CD109 molecule |
| chr1_+_40810516 | 0.22 |
ENST00000435168.2
|
SMAP2
|
small ArfGAP2 |
| chr10_+_51549498 | 0.22 |
ENST00000358559.2
ENST00000298239.6 |
MSMB
|
microseminoprotein, beta- |
| chr1_+_24645865 | 0.22 |
ENST00000342072.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr20_-_7921090 | 0.21 |
ENST00000378789.3
|
HAO1
|
hydroxyacid oxidase (glycolate oxidase) 1 |
| chr13_+_78109804 | 0.21 |
ENST00000535157.1
|
SCEL
|
sciellin |
| chr1_-_11107280 | 0.21 |
ENST00000400897.3
ENST00000400898.3 |
MASP2
|
mannan-binding lectin serine peptidase 2 |
| chrX_+_9431324 | 0.21 |
ENST00000407597.2
ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X
|
transducin (beta)-like 1X-linked |
| chr1_+_24646002 | 0.20 |
ENST00000356046.2
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr1_+_227127981 | 0.20 |
ENST00000366778.1
ENST00000366777.3 ENST00000458507.2 |
ADCK3
|
aarF domain containing kinase 3 |
| chr5_+_95998746 | 0.20 |
ENST00000508608.1
|
CAST
|
calpastatin |
| chr4_+_187148556 | 0.20 |
ENST00000264690.6
ENST00000446598.2 ENST00000414291.1 ENST00000513864.1 |
KLKB1
|
kallikrein B, plasma (Fletcher factor) 1 |
| chr7_+_151791095 | 0.20 |
ENST00000422997.2
|
GALNT11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
| chrX_+_37639302 | 0.20 |
ENST00000545017.1
ENST00000536160.1 |
CYBB
|
cytochrome b-245, beta polypeptide |
| chrX_-_20237059 | 0.19 |
ENST00000457145.1
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| chr2_-_172017393 | 0.19 |
ENST00000442919.2
|
TLK1
|
tousled-like kinase 1 |
| chr6_+_112375462 | 0.19 |
ENST00000361714.1
|
WISP3
|
WNT1 inducible signaling pathway protein 3 |
| chr1_+_35734562 | 0.19 |
ENST00000314607.6
ENST00000373297.2 |
ZMYM4
|
zinc finger, MYM-type 4 |
| chr3_-_186080012 | 0.19 |
ENST00000544847.1
ENST00000265022.3 |
DGKG
|
diacylglycerol kinase, gamma 90kDa |
| chr18_+_60382672 | 0.19 |
ENST00000400316.4
ENST00000262719.5 |
PHLPP1
|
PH domain and leucine rich repeat protein phosphatase 1 |
| chr1_+_61547894 | 0.19 |
ENST00000403491.3
|
NFIA
|
nuclear factor I/A |
| chr2_+_150187020 | 0.18 |
ENST00000334166.4
|
LYPD6
|
LY6/PLAUR domain containing 6 |
| chrX_-_74743080 | 0.18 |
ENST00000373367.3
|
ZDHHC15
|
zinc finger, DHHC-type containing 15 |
| chr7_+_73242490 | 0.18 |
ENST00000431918.1
|
CLDN4
|
claudin 4 |
| chr7_+_134576151 | 0.18 |
ENST00000393118.2
|
CALD1
|
caldesmon 1 |
| chr9_+_116343192 | 0.18 |
ENST00000471324.2
|
RGS3
|
regulator of G-protein signaling 3 |
| chr8_-_30002179 | 0.18 |
ENST00000320542.3
|
MBOAT4
|
membrane bound O-acyltransferase domain containing 4 |
| chr15_+_59910132 | 0.18 |
ENST00000559200.1
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr5_-_135701164 | 0.17 |
ENST00000355180.3
ENST00000426057.2 ENST00000513104.1 |
TRPC7
|
transient receptor potential cation channel, subfamily C, member 7 |
| chr1_+_24645807 | 0.17 |
ENST00000361548.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chrX_+_22050546 | 0.17 |
ENST00000379374.4
|
PHEX
|
phosphate regulating endopeptidase homolog, X-linked |
| chr1_+_145883868 | 0.17 |
ENST00000447947.2
|
GPR89C
|
G protein-coupled receptor 89C |
| chr9_+_6413317 | 0.17 |
ENST00000276893.5
ENST00000381373.3 |
UHRF2
|
ubiquitin-like with PHD and ring finger domains 2, E3 ubiquitin protein ligase |
| chr11_+_119205222 | 0.17 |
ENST00000311413.4
|
RNF26
|
ring finger protein 26 |
| chr2_+_152266392 | 0.16 |
ENST00000444746.2
ENST00000453091.2 ENST00000428287.2 ENST00000433166.2 ENST00000420714.3 ENST00000243326.5 ENST00000414861.2 |
RIF1
|
RAP1 interacting factor homolog (yeast) |
| chr8_-_133772870 | 0.16 |
ENST00000522334.1
ENST00000519016.1 |
TMEM71
|
transmembrane protein 71 |
| chr11_-_26743546 | 0.16 |
ENST00000280467.6
ENST00000396005.3 |
SLC5A12
|
solute carrier family 5 (sodium/monocarboxylate cotransporter), member 12 |
| chr1_+_104159999 | 0.16 |
ENST00000414303.2
ENST00000423678.1 |
AMY2A
|
amylase, alpha 2A (pancreatic) |
| chr1_+_198126209 | 0.16 |
ENST00000367383.1
|
NEK7
|
NIMA-related kinase 7 |
| chr4_-_70080449 | 0.16 |
ENST00000446444.1
|
UGT2B11
|
UDP glucuronosyltransferase 2 family, polypeptide B11 |
| chr11_+_34999328 | 0.16 |
ENST00000526309.1
|
PDHX
|
pyruvate dehydrogenase complex, component X |
| chr1_-_177939348 | 0.15 |
ENST00000464631.2
|
SEC16B
|
SEC16 homolog B (S. cerevisiae) |
| chr2_-_166810261 | 0.15 |
ENST00000243344.7
|
TTC21B
|
tetratricopeptide repeat domain 21B |
| chr4_+_165675269 | 0.15 |
ENST00000507311.1
|
RP11-294O2.2
|
RP11-294O2.2 |
| chr13_-_31038370 | 0.15 |
ENST00000399489.1
ENST00000339872.4 |
HMGB1
|
high mobility group box 1 |
| chr1_+_116654376 | 0.15 |
ENST00000369500.3
|
MAB21L3
|
mab-21-like 3 (C. elegans) |
| chr9_-_98279241 | 0.15 |
ENST00000437951.1
ENST00000375274.2 ENST00000430669.2 ENST00000468211.2 |
PTCH1
|
patched 1 |
| chr12_-_772901 | 0.15 |
ENST00000305108.4
|
NINJ2
|
ninjurin 2 |
| chr19_+_16178317 | 0.15 |
ENST00000344824.6
ENST00000538887.1 |
TPM4
|
tropomyosin 4 |
| chrX_-_30885319 | 0.14 |
ENST00000378933.1
|
TAB3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chr4_-_74486109 | 0.14 |
ENST00000395777.2
|
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr2_-_172290482 | 0.14 |
ENST00000442541.1
ENST00000392599.2 ENST00000375258.4 |
METTL8
|
methyltransferase like 8 |
| chr13_+_76334498 | 0.14 |
ENST00000534657.1
|
LMO7
|
LIM domain 7 |
| chr16_-_87970122 | 0.14 |
ENST00000309893.2
|
CA5A
|
carbonic anhydrase VA, mitochondrial |
| chrX_+_24167828 | 0.14 |
ENST00000379188.3
ENST00000419690.1 ENST00000379177.1 ENST00000304543.5 |
ZFX
|
zinc finger protein, X-linked |
| chr5_+_179921344 | 0.14 |
ENST00000261951.4
|
CNOT6
|
CCR4-NOT transcription complex, subunit 6 |
| chr6_-_165989936 | 0.13 |
ENST00000354448.4
|
PDE10A
|
phosphodiesterase 10A |
| chr14_-_31926623 | 0.13 |
ENST00000356180.4
|
DTD2
|
D-tyrosyl-tRNA deacylase 2 (putative) |
| chr3_+_142315294 | 0.13 |
ENST00000464320.1
|
PLS1
|
plastin 1 |
| chr17_-_27038063 | 0.13 |
ENST00000439862.3
|
PROCA1
|
protein interacting with cyclin A1 |
| chr4_+_124571409 | 0.13 |
ENST00000514823.1
ENST00000511919.1 ENST00000508111.1 |
RP11-93L9.1
|
long intergenic non-protein coding RNA 1091 |
| chr14_+_64971438 | 0.13 |
ENST00000555321.1
|
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr6_+_135502408 | 0.12 |
ENST00000341911.5
ENST00000442647.2 ENST00000316528.8 |
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr5_+_138209688 | 0.12 |
ENST00000518381.1
|
CTNNA1
|
catenin (cadherin-associated protein), alpha 1, 102kDa |
| chr21_+_17442799 | 0.12 |
ENST00000602580.1
ENST00000458468.1 ENST00000602935.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr1_+_200011711 | 0.12 |
ENST00000544748.1
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr18_-_25616519 | 0.12 |
ENST00000399380.3
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chr14_+_56127960 | 0.12 |
ENST00000553624.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr17_+_4613776 | 0.12 |
ENST00000269260.2
|
ARRB2
|
arrestin, beta 2 |
| chr2_-_172017343 | 0.12 |
ENST00000431350.2
ENST00000360843.3 |
TLK1
|
tousled-like kinase 1 |
| chrX_+_37639264 | 0.11 |
ENST00000378588.4
|
CYBB
|
cytochrome b-245, beta polypeptide |
| chr11_+_1151573 | 0.11 |
ENST00000534821.1
ENST00000356191.2 |
MUC5AC
|
mucin 5AC, oligomeric mucus/gel-forming |
| chr5_+_96271141 | 0.11 |
ENST00000231368.5
|
LNPEP
|
leucyl/cystinyl aminopeptidase |
| chr7_-_112579869 | 0.11 |
ENST00000297145.4
|
C7orf60
|
chromosome 7 open reading frame 60 |
| chr12_-_92536433 | 0.11 |
ENST00000551563.2
ENST00000546975.1 ENST00000549802.1 |
C12orf79
|
chromosome 12 open reading frame 79 |
| chr17_+_4613918 | 0.11 |
ENST00000574954.1
ENST00000346341.2 ENST00000572457.1 ENST00000381488.6 ENST00000412477.3 ENST00000571428.1 ENST00000575877.1 |
ARRB2
|
arrestin, beta 2 |
| chr6_+_84222220 | 0.11 |
ENST00000369700.3
|
PRSS35
|
protease, serine, 35 |
| chr4_+_15376165 | 0.11 |
ENST00000382383.3
ENST00000429690.1 |
C1QTNF7
|
C1q and tumor necrosis factor related protein 7 |
| chr7_+_112063192 | 0.10 |
ENST00000005558.4
|
IFRD1
|
interferon-related developmental regulator 1 |
| chr7_-_77325545 | 0.10 |
ENST00000447009.1
ENST00000416650.1 ENST00000440088.1 ENST00000430801.1 ENST00000398043.2 |
RSBN1L-AS1
|
RSBN1L antisense RNA 1 |
| chr2_+_65283506 | 0.10 |
ENST00000377990.2
|
CEP68
|
centrosomal protein 68kDa |
| chr2_+_47630108 | 0.10 |
ENST00000233146.2
ENST00000454849.1 ENST00000543555.1 |
MSH2
|
mutS homolog 2 |
| chr6_+_26251835 | 0.10 |
ENST00000356350.2
|
HIST1H2BH
|
histone cluster 1, H2bh |
| chr1_+_24645915 | 0.09 |
ENST00000350501.5
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr5_-_133510456 | 0.09 |
ENST00000520417.1
|
SKP1
|
S-phase kinase-associated protein 1 |
| chr21_+_17443521 | 0.09 |
ENST00000456342.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr1_+_24646263 | 0.09 |
ENST00000524724.1
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr9_+_140125385 | 0.09 |
ENST00000361134.2
|
SLC34A3
|
solute carrier family 34 (type II sodium/phosphate contransporter), member 3 |
| chr2_+_103089756 | 0.09 |
ENST00000295269.4
|
SLC9A4
|
solute carrier family 9, subfamily A (NHE4, cation proton antiporter 4), member 4 |
| chr2_+_152266604 | 0.09 |
ENST00000430328.2
|
RIF1
|
RAP1 interacting factor homolog (yeast) |
| chr14_-_31926701 | 0.09 |
ENST00000310850.4
|
DTD2
|
D-tyrosyl-tRNA deacylase 2 (putative) |
| chr6_+_7107830 | 0.09 |
ENST00000379933.3
|
RREB1
|
ras responsive element binding protein 1 |
| chr5_+_147774275 | 0.09 |
ENST00000513826.1
|
FBXO38
|
F-box protein 38 |
| chr7_+_77469439 | 0.08 |
ENST00000450574.1
ENST00000416283.2 ENST00000248550.7 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr14_+_56127989 | 0.08 |
ENST00000555573.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr5_-_142065612 | 0.08 |
ENST00000360966.5
ENST00000411960.1 |
FGF1
|
fibroblast growth factor 1 (acidic) |
| chr4_+_170581213 | 0.08 |
ENST00000507875.1
|
CLCN3
|
chloride channel, voltage-sensitive 3 |
| chr5_+_179921430 | 0.08 |
ENST00000393356.1
|
CNOT6
|
CCR4-NOT transcription complex, subunit 6 |
| chrX_+_24167746 | 0.08 |
ENST00000428571.1
ENST00000539115.1 |
ZFX
|
zinc finger protein, X-linked |
| chr21_-_28215332 | 0.08 |
ENST00000517777.1
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr6_-_56707943 | 0.08 |
ENST00000370769.4
ENST00000421834.2 ENST00000312431.6 ENST00000361203.3 ENST00000523817.1 |
DST
|
dystonin |
| chr12_+_78224667 | 0.08 |
ENST00000549464.1
|
NAV3
|
neuron navigator 3 |
| chr7_+_116165754 | 0.07 |
ENST00000405348.1
|
CAV1
|
caveolin 1, caveolae protein, 22kDa |
| chr20_+_31595406 | 0.07 |
ENST00000170150.3
|
BPIFB2
|
BPI fold containing family B, member 2 |
| chr6_-_31550192 | 0.07 |
ENST00000429299.2
ENST00000446745.2 |
LTB
|
lymphotoxin beta (TNF superfamily, member 3) |
| chr6_+_143929307 | 0.07 |
ENST00000427704.2
ENST00000305766.6 |
PHACTR2
|
phosphatase and actin regulator 2 |
| chr6_+_161123270 | 0.07 |
ENST00000366924.2
ENST00000308192.9 ENST00000418964.1 |
PLG
|
plasminogen |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOXA1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:1903615 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.2 | 0.7 | GO:1903565 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.2 | 0.7 | GO:0035623 | renal glucose absorption(GO:0035623) |
| 0.2 | 0.9 | GO:2000174 | regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 0.2 | 1.1 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.2 | 0.8 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.2 | 0.5 | GO:0060730 | regulation of intestinal epithelial structure maintenance(GO:0060730) |
| 0.2 | 1.1 | GO:2000567 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.1 | 0.4 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.1 | 1.0 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 | 0.4 | GO:0060738 | epithelial-mesenchymal signaling involved in prostate gland development(GO:0060738) |
| 0.1 | 0.3 | GO:1902869 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) regulation of amacrine cell differentiation(GO:1902869) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.1 | 0.4 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.1 | 0.5 | GO:0060023 | soft palate development(GO:0060023) |
| 0.1 | 0.3 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.1 | 0.3 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.1 | 0.3 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) |
| 0.1 | 0.3 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.1 | 0.6 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.1 | 0.3 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.1 | 0.4 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.1 | 2.1 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.1 | 0.3 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.1 | 0.3 | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.1 | 0.3 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.0 | 0.2 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.0 | 0.2 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.0 | 0.8 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 1.0 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.4 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.0 | 0.9 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.2 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.1 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.3 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.2 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.0 | 0.8 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.2 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.2 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.4 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.0 | 1.1 | GO:0008211 | glucocorticoid metabolic process(GO:0008211) |
| 0.0 | 0.2 | GO:0002667 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) |
| 0.0 | 0.3 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.2 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.0 | 0.6 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.2 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.3 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.3 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.7 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.0 | 0.3 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.1 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.1 | GO:1900085 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.0 | 0.6 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.0 | 0.4 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.0 | 0.3 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.2 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.0 | 0.4 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.2 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.2 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:2000809 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.0 | 0.4 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.2 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.0 | 0.1 | GO:0060681 | branch elongation involved in ureteric bud branching(GO:0060681) |
| 0.0 | 0.1 | GO:0072502 | cellular phosphate ion homeostasis(GO:0030643) cellular divalent inorganic anion homeostasis(GO:0072501) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.1 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.0 | 0.8 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.0 | 0.2 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 0.3 | GO:0032302 | MutSbeta complex(GO:0032302) |
| 0.1 | 0.4 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.1 | 0.8 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.7 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.3 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.4 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.1 | GO:0070701 | mucus layer(GO:0070701) |
| 0.0 | 0.1 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.0 | 0.3 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.2 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.3 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.2 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 3.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.3 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.1 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.3 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.2 | GO:0016342 | catenin complex(GO:0016342) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.2 | 0.5 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.2 | 2.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.2 | 1.1 | GO:0023030 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.1 | 0.4 | GO:1904713 | beta-catenin destruction complex binding(GO:1904713) |
| 0.1 | 0.3 | GO:0000406 | double-strand/single-strand DNA junction binding(GO:0000406) dinucleotide repeat insertion binding(GO:0032181) |
| 0.1 | 0.3 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.1 | 0.2 | GO:0031859 | platelet activating factor receptor binding(GO:0031859) |
| 0.1 | 0.2 | GO:0051499 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.1 | 0.8 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 0.2 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.1 | 0.2 | GO:0001855 | complement component C4b binding(GO:0001855) |
| 0.1 | 0.2 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.1 | 0.3 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.2 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.2 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 0.0 | 0.2 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 0.2 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 1.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.4 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.3 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.1 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.0 | 0.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:1904854 | proteasome core complex binding(GO:1904854) |
| 0.0 | 0.2 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.9 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.1 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.3 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.2 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 0.8 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.3 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.1 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 0.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 1.2 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 1.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.4 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 2.1 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 1.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.9 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.3 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.7 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.7 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.4 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 1.1 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.5 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.4 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.4 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 1.0 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.1 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |