Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
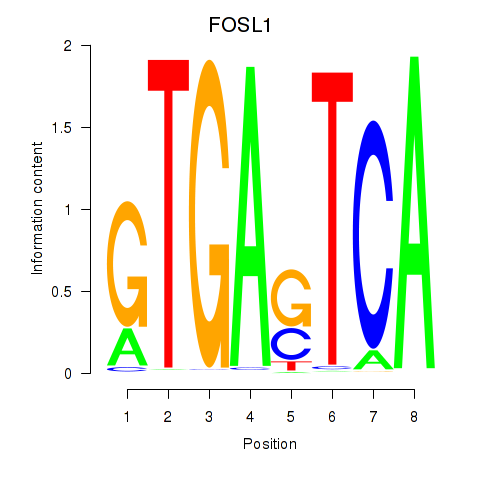
Results for FOSL1
Z-value: 0.45
Transcription factors associated with FOSL1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FOSL1
|
ENSG00000175592.4 | FOS like 1, AP-1 transcription factor subunit |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FOSL1 | hg19_v2_chr11_-_65667997_65668044 | 0.49 | 5.1e-01 | Click! |
Activity profile of FOSL1 motif
Sorted Z-values of FOSL1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_131902346 | 0.41 |
ENST00000432124.1
ENST00000435305.1 |
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr19_-_36019123 | 0.41 |
ENST00000588674.1
ENST00000452271.2 ENST00000518157.1 |
SBSN
|
suprabasin |
| chr19_-_19739321 | 0.29 |
ENST00000588461.1
|
LPAR2
|
lysophosphatidic acid receptor 2 |
| chr20_-_44455976 | 0.27 |
ENST00000372555.3
|
TNNC2
|
troponin C type 2 (fast) |
| chr19_-_35981358 | 0.26 |
ENST00000484218.2
ENST00000338897.3 |
KRTDAP
|
keratinocyte differentiation-associated protein |
| chr17_-_39769005 | 0.24 |
ENST00000301653.4
ENST00000593067.1 |
KRT16
|
keratin 16 |
| chr1_-_12679171 | 0.21 |
ENST00000606790.1
|
RP11-474O21.5
|
RP11-474O21.5 |
| chr3_+_57094469 | 0.20 |
ENST00000334325.1
|
SPATA12
|
spermatogenesis associated 12 |
| chr2_+_27505260 | 0.19 |
ENST00000380075.2
ENST00000296098.4 |
TRIM54
|
tripartite motif containing 54 |
| chr8_-_123139423 | 0.18 |
ENST00000523792.1
|
RP11-398G24.2
|
RP11-398G24.2 |
| chr2_+_87755054 | 0.18 |
ENST00000423846.1
|
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr19_+_19516561 | 0.18 |
ENST00000457895.2
|
GATAD2A
|
GATA zinc finger domain containing 2A |
| chr15_+_57211318 | 0.17 |
ENST00000557947.1
|
TCF12
|
transcription factor 12 |
| chr16_-_30997533 | 0.16 |
ENST00000602217.1
|
AC135048.1
|
Uncharacterized protein |
| chr20_+_44637526 | 0.16 |
ENST00000372330.3
|
MMP9
|
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) |
| chr7_+_33765593 | 0.16 |
ENST00000311067.3
|
RP11-89N17.1
|
HCG1643653; Uncharacterized protein |
| chr10_+_1102303 | 0.16 |
ENST00000381329.1
|
WDR37
|
WD repeat domain 37 |
| chr3_+_184534994 | 0.16 |
ENST00000441141.1
ENST00000445089.1 |
VPS8
|
vacuolar protein sorting 8 homolog (S. cerevisiae) |
| chr15_-_44486632 | 0.15 |
ENST00000484674.1
|
FRMD5
|
FERM domain containing 5 |
| chr11_-_66103932 | 0.15 |
ENST00000311320.4
|
RIN1
|
Ras and Rab interactor 1 |
| chr9_-_127177703 | 0.14 |
ENST00000259457.3
ENST00000536392.1 ENST00000441097.1 |
PSMB7
|
proteasome (prosome, macropain) subunit, beta type, 7 |
| chr15_+_75335604 | 0.14 |
ENST00000563393.1
|
PPCDC
|
phosphopantothenoylcysteine decarboxylase |
| chr1_+_156096336 | 0.14 |
ENST00000504687.1
ENST00000473598.2 |
LMNA
|
lamin A/C |
| chr9_-_34662651 | 0.14 |
ENST00000259631.4
|
CCL27
|
chemokine (C-C motif) ligand 27 |
| chr2_-_61389240 | 0.14 |
ENST00000606876.1
|
RP11-493E12.1
|
RP11-493E12.1 |
| chr2_+_87808725 | 0.13 |
ENST00000413202.1
|
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr18_-_74207146 | 0.13 |
ENST00000443185.2
|
ZNF516
|
zinc finger protein 516 |
| chr19_-_6057282 | 0.13 |
ENST00000592281.1
|
RFX2
|
regulatory factor X, 2 (influences HLA class II expression) |
| chr17_+_28256874 | 0.13 |
ENST00000541045.1
ENST00000536908.2 |
EFCAB5
|
EF-hand calcium binding domain 5 |
| chr6_-_35888858 | 0.13 |
ENST00000507909.1
|
SRPK1
|
SRSF protein kinase 1 |
| chr3_-_18466787 | 0.12 |
ENST00000338745.6
ENST00000450898.1 |
SATB1
|
SATB homeobox 1 |
| chr14_+_77425972 | 0.12 |
ENST00000553613.1
|
RP11-7F17.7
|
RP11-7F17.7 |
| chr11_-_66104237 | 0.12 |
ENST00000530056.1
|
RIN1
|
Ras and Rab interactor 1 |
| chr1_-_45272951 | 0.12 |
ENST00000372200.1
|
TCTEX1D4
|
Tctex1 domain containing 4 |
| chr10_+_88780049 | 0.12 |
ENST00000343959.4
|
FAM25A
|
family with sequence similarity 25, member A |
| chr12_+_10365082 | 0.12 |
ENST00000545859.1
|
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr1_+_17634689 | 0.12 |
ENST00000375453.1
ENST00000375448.4 |
PADI4
|
peptidyl arginine deiminase, type IV |
| chr7_+_102191679 | 0.12 |
ENST00000507918.1
ENST00000432940.1 |
SPDYE2
|
speedy/RINGO cell cycle regulator family member E2 |
| chrX_+_37765364 | 0.11 |
ENST00000567273.1
|
AL121578.2
|
AL121578.2 |
| chr18_-_68004529 | 0.11 |
ENST00000578633.1
|
RP11-484N16.1
|
RP11-484N16.1 |
| chr17_-_28257080 | 0.11 |
ENST00000579954.1
ENST00000540801.1 ENST00000269033.3 ENST00000590153.1 ENST00000582084.1 |
SSH2
|
slingshot protein phosphatase 2 |
| chr10_+_115312766 | 0.11 |
ENST00000351270.3
|
HABP2
|
hyaluronan binding protein 2 |
| chr12_-_719573 | 0.11 |
ENST00000397265.3
|
NINJ2
|
ninjurin 2 |
| chr3_-_196065248 | 0.11 |
ENST00000446879.1
ENST00000273695.3 |
TM4SF19
|
transmembrane 4 L six family member 19 |
| chr16_-_74330612 | 0.11 |
ENST00000569389.1
ENST00000562888.1 |
AC009120.4
|
AC009120.4 |
| chr8_-_68658578 | 0.11 |
ENST00000518549.1
ENST00000297770.4 ENST00000297769.4 |
CPA6
|
carboxypeptidase A6 |
| chr10_-_31288398 | 0.10 |
ENST00000538351.2
|
ZNF438
|
zinc finger protein 438 |
| chr3_+_112930387 | 0.10 |
ENST00000485230.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr7_-_96339132 | 0.10 |
ENST00000413065.1
|
SHFM1
|
split hand/foot malformation (ectrodactyly) type 1 |
| chr11_-_47207390 | 0.10 |
ENST00000539589.1
ENST00000528462.1 |
PACSIN3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr1_+_150122034 | 0.10 |
ENST00000025469.6
ENST00000369124.4 |
PLEKHO1
|
pleckstrin homology domain containing, family O member 1 |
| chr1_+_27189631 | 0.10 |
ENST00000339276.4
|
SFN
|
stratifin |
| chr16_+_28875126 | 0.10 |
ENST00000359285.5
ENST00000538342.1 |
SH2B1
|
SH2B adaptor protein 1 |
| chr11_-_2924720 | 0.10 |
ENST00000455942.2
|
SLC22A18AS
|
solute carrier family 22 (organic cation transporter), member 18 antisense |
| chr11_+_394196 | 0.10 |
ENST00000331563.2
ENST00000531857.1 |
PKP3
|
plakophilin 3 |
| chr11_+_36397915 | 0.10 |
ENST00000526682.1
ENST00000530252.1 |
PRR5L
|
proline rich 5 like |
| chr16_+_2083265 | 0.10 |
ENST00000565855.1
ENST00000566198.1 |
SLC9A3R2
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 2 |
| chr19_+_50431959 | 0.10 |
ENST00000595125.1
|
ATF5
|
activating transcription factor 5 |
| chr22_+_22723969 | 0.10 |
ENST00000390295.2
|
IGLV7-46
|
immunoglobulin lambda variable 7-46 (gene/pseudogene) |
| chr17_-_42994283 | 0.09 |
ENST00000593179.1
|
GFAP
|
glial fibrillary acidic protein |
| chr11_+_82783097 | 0.09 |
ENST00000501011.2
ENST00000527627.1 ENST00000526795.1 ENST00000533528.1 ENST00000533708.1 ENST00000534499.1 |
RAB30-AS1
|
RAB30 antisense RNA 1 (head to head) |
| chr10_-_6019552 | 0.09 |
ENST00000379977.3
ENST00000397251.3 ENST00000397248.2 |
IL15RA
|
interleukin 15 receptor, alpha |
| chr12_+_57914742 | 0.09 |
ENST00000551351.1
|
MBD6
|
methyl-CpG binding domain protein 6 |
| chr5_-_141061777 | 0.09 |
ENST00000239440.4
|
ARAP3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
| chr14_+_69726864 | 0.09 |
ENST00000448469.3
|
GALNT16
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 16 |
| chr21_+_30672433 | 0.09 |
ENST00000451655.1
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chr5_+_35856951 | 0.09 |
ENST00000303115.3
ENST00000343305.4 ENST00000506850.1 ENST00000511982.1 |
IL7R
|
interleukin 7 receptor |
| chr9_+_140135665 | 0.09 |
ENST00000340384.4
|
TUBB4B
|
tubulin, beta 4B class IVb |
| chr11_-_64013288 | 0.09 |
ENST00000542235.1
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr2_-_224467002 | 0.09 |
ENST00000421386.1
ENST00000433889.1 |
SCG2
|
secretogranin II |
| chr17_+_7590734 | 0.09 |
ENST00000457584.2
|
WRAP53
|
WD repeat containing, antisense to TP53 |
| chr6_-_44233361 | 0.09 |
ENST00000275015.5
|
NFKBIE
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon |
| chr1_+_156084461 | 0.09 |
ENST00000347559.2
ENST00000361308.4 ENST00000368300.4 ENST00000368299.3 |
LMNA
|
lamin A/C |
| chr17_+_49230897 | 0.09 |
ENST00000393196.3
ENST00000336097.3 ENST00000480143.1 ENST00000511355.1 ENST00000013034.3 ENST00000393198.3 ENST00000608447.1 ENST00000393193.2 ENST00000376392.6 ENST00000555572.1 |
NME1
NME1-NME2
NME2
|
NME/NM23 nucleoside diphosphate kinase 1 NME1-NME2 readthrough NME/NM23 nucleoside diphosphate kinase 2 |
| chr12_-_56236711 | 0.09 |
ENST00000409200.3
|
MMP19
|
matrix metallopeptidase 19 |
| chr5_-_141061759 | 0.09 |
ENST00000508305.1
|
ARAP3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
| chr12_+_56546223 | 0.09 |
ENST00000550443.1
ENST00000207437.5 |
MYL6B
|
myosin, light chain 6B, alkali, smooth muscle and non-muscle |
| chr17_-_73781567 | 0.09 |
ENST00000586607.1
|
H3F3B
|
H3 histone, family 3B (H3.3B) |
| chr1_+_156095951 | 0.08 |
ENST00000448611.2
ENST00000368297.1 |
LMNA
|
lamin A/C |
| chr15_+_91416092 | 0.08 |
ENST00000559353.1
|
FURIN
|
furin (paired basic amino acid cleaving enzyme) |
| chr3_-_196065374 | 0.08 |
ENST00000454715.1
|
TM4SF19
|
transmembrane 4 L six family member 19 |
| chr10_-_81708854 | 0.08 |
ENST00000372292.3
|
SFTPD
|
surfactant protein D |
| chr7_-_42276612 | 0.08 |
ENST00000395925.3
ENST00000437480.1 |
GLI3
|
GLI family zinc finger 3 |
| chr12_+_56546363 | 0.08 |
ENST00000551834.1
ENST00000552568.1 |
MYL6B
|
myosin, light chain 6B, alkali, smooth muscle and non-muscle |
| chr19_-_40931891 | 0.08 |
ENST00000357949.4
|
SERTAD1
|
SERTA domain containing 1 |
| chr11_-_6341844 | 0.08 |
ENST00000303927.3
|
PRKCDBP
|
protein kinase C, delta binding protein |
| chr15_-_78112553 | 0.08 |
ENST00000562933.1
|
LINGO1
|
leucine rich repeat and Ig domain containing 1 |
| chr11_-_102826434 | 0.08 |
ENST00000340273.4
ENST00000260302.3 |
MMP13
|
matrix metallopeptidase 13 (collagenase 3) |
| chr3_-_129513259 | 0.08 |
ENST00000329333.5
|
TMCC1
|
transmembrane and coiled-coil domain family 1 |
| chr1_+_39670360 | 0.08 |
ENST00000494012.1
|
MACF1
|
microtubule-actin crosslinking factor 1 |
| chr2_-_220173685 | 0.08 |
ENST00000423636.2
ENST00000442029.1 ENST00000412847.1 |
PTPRN
|
protein tyrosine phosphatase, receptor type, N |
| chrX_-_153285251 | 0.08 |
ENST00000444230.1
ENST00000393682.1 ENST00000393687.2 ENST00000429936.2 ENST00000369974.2 |
IRAK1
|
interleukin-1 receptor-associated kinase 1 |
| chr22_+_37959647 | 0.08 |
ENST00000415670.1
|
CDC42EP1
|
CDC42 effector protein (Rho GTPase binding) 1 |
| chr15_-_72521017 | 0.08 |
ENST00000561609.1
|
PKM
|
pyruvate kinase, muscle |
| chr19_+_36203830 | 0.08 |
ENST00000262630.3
|
ZBTB32
|
zinc finger and BTB domain containing 32 |
| chr3_+_174158732 | 0.08 |
ENST00000434257.1
|
NAALADL2
|
N-acetylated alpha-linked acidic dipeptidase-like 2 |
| chr7_-_96339167 | 0.08 |
ENST00000444799.1
ENST00000417009.1 ENST00000248566.2 |
SHFM1
|
split hand/foot malformation (ectrodactyly) type 1 |
| chr12_-_102591604 | 0.08 |
ENST00000329406.4
|
PMCH
|
pro-melanin-concentrating hormone |
| chr20_+_48909240 | 0.08 |
ENST00000371639.3
|
RP11-290F20.1
|
RP11-290F20.1 |
| chr17_-_27045405 | 0.08 |
ENST00000430132.2
|
RAB34
|
RAB34, member RAS oncogene family |
| chrX_-_153285395 | 0.08 |
ENST00000369980.3
|
IRAK1
|
interleukin-1 receptor-associated kinase 1 |
| chr19_+_50432400 | 0.08 |
ENST00000423777.2
ENST00000600336.1 ENST00000597227.1 |
ATF5
|
activating transcription factor 5 |
| chr17_-_18908040 | 0.08 |
ENST00000388995.6
|
FAM83G
|
family with sequence similarity 83, member G |
| chr2_-_220083076 | 0.08 |
ENST00000295750.4
|
ABCB6
|
ATP-binding cassette, sub-family B (MDR/TAP), member 6 |
| chr12_-_53297432 | 0.07 |
ENST00000546900.1
|
KRT8
|
keratin 8 |
| chr18_+_55888767 | 0.07 |
ENST00000431212.2
ENST00000586268.1 ENST00000587190.1 |
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr2_-_220117867 | 0.07 |
ENST00000456818.1
ENST00000447205.1 |
TUBA4A
|
tubulin, alpha 4a |
| chr11_-_62323702 | 0.07 |
ENST00000530285.1
|
AHNAK
|
AHNAK nucleoprotein |
| chr4_+_76481258 | 0.07 |
ENST00000311623.4
ENST00000435974.2 |
C4orf26
|
chromosome 4 open reading frame 26 |
| chr1_+_24645807 | 0.07 |
ENST00000361548.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr14_+_63671105 | 0.07 |
ENST00000316754.3
|
RHOJ
|
ras homolog family member J |
| chr16_-_4665023 | 0.07 |
ENST00000591897.1
|
UBALD1
|
UBA-like domain containing 1 |
| chr16_+_28875268 | 0.07 |
ENST00000395532.4
|
SH2B1
|
SH2B adaptor protein 1 |
| chr1_-_113247543 | 0.07 |
ENST00000414971.1
ENST00000534717.1 |
RHOC
|
ras homolog family member C |
| chr19_-_44285401 | 0.07 |
ENST00000262888.3
|
KCNN4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
| chr21_+_30673091 | 0.07 |
ENST00000447177.1
|
BACH1
|
BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
| chr6_+_25652501 | 0.07 |
ENST00000334979.6
|
SCGN
|
secretagogin, EF-hand calcium binding protein |
| chr20_+_48429233 | 0.07 |
ENST00000417961.1
|
SLC9A8
|
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chr21_+_27543175 | 0.07 |
ENST00000608591.1
ENST00000609365.1 |
AP000230.1
|
AP000230.1 |
| chr11_-_64013663 | 0.07 |
ENST00000392210.2
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr2_-_142888573 | 0.07 |
ENST00000434794.1
|
LRP1B
|
low density lipoprotein receptor-related protein 1B |
| chr11_+_34643600 | 0.07 |
ENST00000530286.1
ENST00000533754.1 |
EHF
|
ets homologous factor |
| chrX_-_48901012 | 0.07 |
ENST00000315869.7
|
TFE3
|
transcription factor binding to IGHM enhancer 3 |
| chr12_-_53625958 | 0.07 |
ENST00000327550.3
ENST00000546717.1 ENST00000425354.2 ENST00000394426.1 |
RARG
|
retinoic acid receptor, gamma |
| chr17_-_74351010 | 0.07 |
ENST00000435555.2
|
PRPSAP1
|
phosphoribosyl pyrophosphate synthetase-associated protein 1 |
| chr12_-_57914275 | 0.07 |
ENST00000547303.1
ENST00000552740.1 ENST00000547526.1 ENST00000551116.1 ENST00000346473.3 |
DDIT3
|
DNA-damage-inducible transcript 3 |
| chr5_-_41794313 | 0.07 |
ENST00000512084.1
|
OXCT1
|
3-oxoacid CoA transferase 1 |
| chr5_-_138534071 | 0.07 |
ENST00000394817.2
|
SIL1
|
SIL1 nucleotide exchange factor |
| chr12_-_56122426 | 0.07 |
ENST00000551173.1
|
CD63
|
CD63 molecule |
| chr12_-_56122761 | 0.06 |
ENST00000552164.1
ENST00000420846.3 ENST00000257857.4 |
CD63
|
CD63 molecule |
| chr1_+_223889310 | 0.06 |
ENST00000434648.1
|
CAPN2
|
calpain 2, (m/II) large subunit |
| chr6_-_30685214 | 0.06 |
ENST00000425072.1
|
MDC1
|
mediator of DNA-damage checkpoint 1 |
| chr1_+_24645915 | 0.06 |
ENST00000350501.5
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr9_-_86322516 | 0.06 |
ENST00000529923.1
|
UBQLN1
|
ubiquilin 1 |
| chr12_-_53343602 | 0.06 |
ENST00000546897.1
ENST00000552551.1 |
KRT8
|
keratin 8 |
| chr16_-_1429627 | 0.06 |
ENST00000248104.7
|
UNKL
|
unkempt family zinc finger-like |
| chr16_+_30968615 | 0.06 |
ENST00000262519.8
|
SETD1A
|
SET domain containing 1A |
| chr16_+_83986827 | 0.06 |
ENST00000393306.1
ENST00000565123.1 |
OSGIN1
|
oxidative stress induced growth inhibitor 1 |
| chr17_-_27405875 | 0.06 |
ENST00000359450.6
|
TIAF1
|
TGFB1-induced anti-apoptotic factor 1 |
| chr16_+_30418910 | 0.06 |
ENST00000566625.1
|
ZNF771
|
zinc finger protein 771 |
| chr1_+_24645865 | 0.06 |
ENST00000342072.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr1_+_24646002 | 0.06 |
ENST00000356046.2
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr19_-_4559814 | 0.06 |
ENST00000586582.1
|
SEMA6B
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6B |
| chr3_+_112930946 | 0.06 |
ENST00000462425.1
|
BOC
|
BOC cell adhesion associated, oncogene regulated |
| chr8_+_120220561 | 0.06 |
ENST00000276681.6
|
MAL2
|
mal, T-cell differentiation protein 2 (gene/pseudogene) |
| chr5_-_41794663 | 0.06 |
ENST00000510634.1
|
OXCT1
|
3-oxoacid CoA transferase 1 |
| chr9_-_130341268 | 0.06 |
ENST00000373314.3
|
FAM129B
|
family with sequence similarity 129, member B |
| chr11_-_119066545 | 0.06 |
ENST00000415318.1
|
CCDC153
|
coiled-coil domain containing 153 |
| chr2_+_135596180 | 0.06 |
ENST00000283054.4
ENST00000392928.1 |
ACMSD
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr16_+_57680043 | 0.06 |
ENST00000569154.1
|
GPR56
|
G protein-coupled receptor 56 |
| chr1_+_151227179 | 0.06 |
ENST00000368884.3
ENST00000368881.4 |
PSMD4
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 4 |
| chr1_+_36621529 | 0.06 |
ENST00000316156.4
|
MAP7D1
|
MAP7 domain containing 1 |
| chr16_-_72206034 | 0.06 |
ENST00000537465.1
ENST00000237353.10 |
PMFBP1
|
polyamine modulated factor 1 binding protein 1 |
| chr11_+_35639735 | 0.06 |
ENST00000317811.4
|
FJX1
|
four jointed box 1 (Drosophila) |
| chr7_-_150721570 | 0.06 |
ENST00000377974.2
ENST00000444312.1 ENST00000605938.1 ENST00000605952.1 |
ATG9B
|
autophagy related 9B |
| chr12_+_52695617 | 0.06 |
ENST00000293525.5
|
KRT86
|
keratin 86 |
| chr2_-_166651191 | 0.06 |
ENST00000392701.3
|
GALNT3
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3) |
| chr3_+_183903811 | 0.06 |
ENST00000429586.2
ENST00000292808.5 |
ABCF3
|
ATP-binding cassette, sub-family F (GCN20), member 3 |
| chr1_-_159894319 | 0.06 |
ENST00000320307.4
|
TAGLN2
|
transgelin 2 |
| chr1_+_33722080 | 0.06 |
ENST00000483388.1
ENST00000539719.1 |
ZNF362
|
zinc finger protein 362 |
| chr16_+_30077098 | 0.06 |
ENST00000395240.3
ENST00000566846.1 |
ALDOA
|
aldolase A, fructose-bisphosphate |
| chrX_-_129244336 | 0.06 |
ENST00000434609.1
|
ELF4
|
E74-like factor 4 (ets domain transcription factor) |
| chr6_-_31704282 | 0.06 |
ENST00000375784.3
ENST00000375779.2 |
CLIC1
|
chloride intracellular channel 1 |
| chr4_+_3388057 | 0.06 |
ENST00000538395.1
|
RGS12
|
regulator of G-protein signaling 12 |
| chr8_-_27472198 | 0.06 |
ENST00000519472.1
ENST00000523589.1 ENST00000522413.1 ENST00000523396.1 ENST00000560366.1 |
CLU
|
clusterin |
| chr11_-_57417367 | 0.05 |
ENST00000534810.1
|
YPEL4
|
yippee-like 4 (Drosophila) |
| chr15_-_67439270 | 0.05 |
ENST00000558463.1
|
RP11-342M21.2
|
Uncharacterized protein |
| chr12_+_56732658 | 0.05 |
ENST00000228534.4
|
IL23A
|
interleukin 23, alpha subunit p19 |
| chr5_+_177540444 | 0.05 |
ENST00000274605.5
|
N4BP3
|
NEDD4 binding protein 3 |
| chr10_-_6019984 | 0.05 |
ENST00000525219.2
|
IL15RA
|
interleukin 15 receptor, alpha |
| chr5_-_137071689 | 0.05 |
ENST00000505853.1
|
KLHL3
|
kelch-like family member 3 |
| chr12_+_6644443 | 0.05 |
ENST00000396858.1
|
GAPDH
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr1_-_160990886 | 0.05 |
ENST00000537746.1
|
F11R
|
F11 receptor |
| chr1_-_151965048 | 0.05 |
ENST00000368809.1
|
S100A10
|
S100 calcium binding protein A10 |
| chr16_-_1429674 | 0.05 |
ENST00000403703.1
ENST00000397464.1 ENST00000402641.2 |
UNKL
|
unkempt family zinc finger-like |
| chr1_+_36621697 | 0.05 |
ENST00000373150.4
ENST00000373151.2 |
MAP7D1
|
MAP7 domain containing 1 |
| chr17_+_73780852 | 0.05 |
ENST00000589666.1
|
UNK
|
unkempt family zinc finger |
| chr19_+_18485509 | 0.05 |
ENST00000597765.1
|
GDF15
|
growth differentiation factor 15 |
| chr19_+_50706866 | 0.05 |
ENST00000440075.2
ENST00000376970.2 ENST00000425460.1 ENST00000599920.1 ENST00000601313.1 |
MYH14
|
myosin, heavy chain 14, non-muscle |
| chr21_+_26934165 | 0.05 |
ENST00000456917.1
|
MIR155HG
|
MIR155 host gene (non-protein coding) |
| chrX_+_153769446 | 0.05 |
ENST00000422680.1
|
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr18_+_74207477 | 0.05 |
ENST00000532511.1
|
RP11-17M16.1
|
uncharacterized protein LOC400658 |
| chr1_-_158656488 | 0.05 |
ENST00000368147.4
|
SPTA1
|
spectrin, alpha, erythrocytic 1 (elliptocytosis 2) |
| chr17_+_74381343 | 0.05 |
ENST00000392496.3
|
SPHK1
|
sphingosine kinase 1 |
| chr11_+_118958689 | 0.05 |
ENST00000535253.1
ENST00000392841.1 |
HMBS
|
hydroxymethylbilane synthase |
| chr5_-_141030943 | 0.05 |
ENST00000522783.1
ENST00000519800.1 ENST00000435817.2 |
FCHSD1
|
FCH and double SH3 domains 1 |
| chr1_-_179112189 | 0.05 |
ENST00000512653.1
ENST00000344730.3 |
ABL2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr15_-_70388599 | 0.05 |
ENST00000560996.1
ENST00000558201.1 |
TLE3
|
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) |
| chr19_-_10450328 | 0.05 |
ENST00000160262.5
|
ICAM3
|
intercellular adhesion molecule 3 |
| chr5_+_140729649 | 0.05 |
ENST00000523390.1
|
PCDHGB1
|
protocadherin gamma subfamily B, 1 |
| chr8_-_67974552 | 0.05 |
ENST00000357849.4
|
COPS5
|
COP9 signalosome subunit 5 |
| chr18_-_25616519 | 0.05 |
ENST00000399380.3
|
CDH2
|
cadherin 2, type 1, N-cadherin (neuronal) |
| chr16_-_4664860 | 0.05 |
ENST00000587615.1
ENST00000587649.1 ENST00000590965.1 ENST00000591401.1 ENST00000283474.7 |
UBALD1
|
UBA-like domain containing 1 |
| chr2_-_166651152 | 0.05 |
ENST00000431484.1
ENST00000412248.1 |
GALNT3
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3) |
| chr16_+_57662419 | 0.05 |
ENST00000388812.4
ENST00000538815.1 ENST00000456916.1 ENST00000567154.1 ENST00000388813.5 ENST00000562558.1 ENST00000566271.2 |
GPR56
|
G protein-coupled receptor 56 |
| chr11_-_65667884 | 0.05 |
ENST00000448083.2
ENST00000531493.1 ENST00000532401.1 |
FOSL1
|
FOS-like antigen 1 |
| chr5_-_138533973 | 0.05 |
ENST00000507002.1
ENST00000505830.1 ENST00000508639.1 ENST00000265195.5 |
SIL1
|
SIL1 nucleotide exchange factor |
| chr17_+_28268623 | 0.05 |
ENST00000394835.3
ENST00000320856.5 ENST00000394832.2 ENST00000378738.3 |
EFCAB5
|
EF-hand calcium binding domain 5 |
| chr20_+_48429356 | 0.05 |
ENST00000361573.2
ENST00000541138.1 ENST00000539601.1 |
SLC9A8
|
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chr17_+_48712138 | 0.05 |
ENST00000515707.1
|
ABCC3
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of FOSL1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.0 | 0.3 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.1 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.0 | 0.1 | GO:0090472 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.0 | 0.1 | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.0 | 0.1 | GO:0006117 | acetaldehyde metabolic process(GO:0006117) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.1 | GO:1901340 | negative regulation of store-operated calcium channel activity(GO:1901340) |
| 0.0 | 0.4 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 0.0 | 0.1 | GO:0044028 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.0 | 0.1 | GO:0010482 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.0 | 0.1 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.0 | 0.1 | GO:0001554 | luteolysis(GO:0001554) |
| 0.0 | 0.1 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 | 0.2 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.1 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.1 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.0 | 0.1 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.0 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.0 | 0.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 | 0.2 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.0 | 0.1 | GO:0045085 | negative regulation of interleukin-2 biosynthetic process(GO:0045085) |
| 0.0 | 0.1 | GO:0072156 | distal tubule morphogenesis(GO:0072156) |
| 0.0 | 0.0 | GO:0030264 | nuclear fragmentation involved in apoptotic nuclear change(GO:0030264) |
| 0.0 | 0.3 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.2 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.0 | 0.0 | GO:0071623 | negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.0 | 0.0 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.1 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.0 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.1 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.2 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.0 | 0.0 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:0070384 | Harderian gland development(GO:0070384) |
| 0.0 | 0.0 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.0 | 0.2 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 0.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.2 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.1 | GO:0035525 | NF-kappaB p50/p65 complex(GO:0035525) |
| 0.0 | 0.1 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.2 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.4 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.2 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.1 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.0 | 0.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.0 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.0 | 0.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.0 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.0 | 0.1 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 0.3 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.1 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.0 | 0.1 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.2 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.1 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.0 | 0.0 | GO:0015432 | bile acid-exporting ATPase activity(GO:0015432) |
| 0.0 | 0.0 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.0 | 0.0 | GO:0003955 | NAD(P)H dehydrogenase (quinone) activity(GO:0003955) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) aspartic-type endopeptidase inhibitor activity(GO:0019828) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.0 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.0 | 0.1 | GO:1990450 | linear polyubiquitin binding(GO:1990450) |
| 0.0 | 0.0 | GO:0005019 | platelet-derived growth factor beta-receptor activity(GO:0005019) |
| 0.0 | 0.0 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.0 | 0.0 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.0 | 0.0 | GO:0005151 | interleukin-1, Type II receptor binding(GO:0005151) |
| 0.0 | 0.0 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |