Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
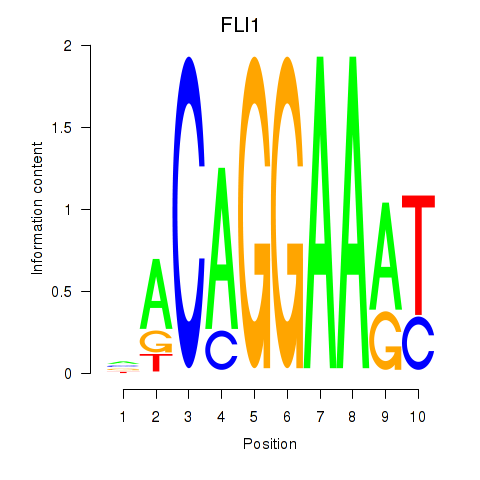
Results for FLI1
Z-value: 0.40
Transcription factors associated with FLI1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FLI1
|
ENSG00000151702.12 | Fli-1 proto-oncogene, ETS transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| FLI1 | hg19_v2_chr11_+_128562372_128562397 | 0.83 | 1.7e-01 | Click! |
Activity profile of FLI1 motif
Sorted Z-values of FLI1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_119316721 | 0.29 |
ENST00000488919.1
ENST00000495992.1 |
PLA1A
|
phospholipase A1 member A |
| chr8_+_74903580 | 0.24 |
ENST00000284818.2
ENST00000518893.1 |
LY96
|
lymphocyte antigen 96 |
| chr3_+_119316689 | 0.23 |
ENST00000273371.4
|
PLA1A
|
phospholipase A1 member A |
| chr1_-_63988846 | 0.23 |
ENST00000283568.8
ENST00000371092.3 ENST00000271002.10 |
ITGB3BP
|
integrin beta 3 binding protein (beta3-endonexin) |
| chr11_-_9482010 | 0.20 |
ENST00000596206.1
|
AC132192.1
|
LOC644656 protein; Uncharacterized protein |
| chr17_-_7082861 | 0.20 |
ENST00000269299.3
|
ASGR1
|
asialoglycoprotein receptor 1 |
| chr14_+_32414059 | 0.19 |
ENST00000553330.1
|
RP11-187E13.1
|
Uncharacterized protein |
| chr1_+_229440129 | 0.18 |
ENST00000366688.3
|
SPHAR
|
S-phase response (cyclin related) |
| chr12_+_97300995 | 0.17 |
ENST00000266742.4
ENST00000429527.2 ENST00000554226.1 ENST00000557478.1 ENST00000557092.1 ENST00000411739.2 ENST00000553609.1 |
NEDD1
|
neural precursor cell expressed, developmentally down-regulated 1 |
| chr3_+_52321827 | 0.17 |
ENST00000473032.1
ENST00000305690.8 ENST00000354773.4 ENST00000471180.1 ENST00000436784.2 |
GLYCTK
|
glycerate kinase |
| chr4_+_20702059 | 0.16 |
ENST00000444671.2
ENST00000510700.1 ENST00000506745.1 ENST00000514663.1 ENST00000509469.1 ENST00000515339.1 ENST00000513861.1 ENST00000502374.1 ENST00000538990.1 ENST00000511160.1 ENST00000504630.1 ENST00000513590.1 ENST00000514292.1 ENST00000502938.1 ENST00000509625.1 ENST00000505160.1 ENST00000507634.1 ENST00000513459.1 ENST00000511089.1 |
PACRGL
|
PARK2 co-regulated-like |
| chr2_+_198570081 | 0.15 |
ENST00000282276.6
|
MARS2
|
methionyl-tRNA synthetase 2, mitochondrial |
| chr4_+_89300158 | 0.15 |
ENST00000502870.1
|
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr19_+_16296191 | 0.15 |
ENST00000589852.1
ENST00000263384.7 ENST00000588367.1 ENST00000587351.1 |
FAM32A
|
family with sequence similarity 32, member A |
| chr16_-_20817753 | 0.15 |
ENST00000389345.5
ENST00000300005.3 ENST00000357967.4 ENST00000569729.1 |
ERI2
|
ERI1 exoribonuclease family member 2 |
| chr1_+_59486129 | 0.15 |
ENST00000438195.1
ENST00000424308.1 |
RP4-794H19.4
|
RP4-794H19.4 |
| chr1_-_161039647 | 0.15 |
ENST00000368013.3
|
ARHGAP30
|
Rho GTPase activating protein 30 |
| chr12_+_94071129 | 0.15 |
ENST00000552983.1
ENST00000332896.3 ENST00000552033.1 ENST00000548483.1 |
CRADD
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chrX_+_70435044 | 0.14 |
ENST00000374029.1
ENST00000374022.3 ENST00000447581.1 |
GJB1
|
gap junction protein, beta 1, 32kDa |
| chr4_-_155533787 | 0.14 |
ENST00000407946.1
ENST00000405164.1 ENST00000336098.3 ENST00000393846.2 ENST00000404648.3 ENST00000443553.1 |
FGG
|
fibrinogen gamma chain |
| chr1_+_18807424 | 0.14 |
ENST00000400664.1
|
KLHDC7A
|
kelch domain containing 7A |
| chr9_-_75567962 | 0.14 |
ENST00000297785.3
ENST00000376939.1 |
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1 |
| chr11_-_67205538 | 0.13 |
ENST00000326294.3
|
PTPRCAP
|
protein tyrosine phosphatase, receptor type, C-associated protein |
| chr3_-_195310802 | 0.13 |
ENST00000421243.1
ENST00000453131.1 |
APOD
|
apolipoprotein D |
| chr14_+_100531615 | 0.13 |
ENST00000392920.3
|
EVL
|
Enah/Vasp-like |
| chr16_+_20775024 | 0.13 |
ENST00000289416.5
|
ACSM3
|
acyl-CoA synthetase medium-chain family member 3 |
| chr19_-_36297632 | 0.13 |
ENST00000588266.2
|
PRODH2
|
proline dehydrogenase (oxidase) 2 |
| chr7_+_2281843 | 0.13 |
ENST00000356714.1
ENST00000397049.1 |
NUDT1
|
nudix (nucleoside diphosphate linked moiety X)-type motif 1 |
| chr6_+_125474795 | 0.13 |
ENST00000304877.13
ENST00000534000.1 ENST00000368402.5 ENST00000368388.2 |
TPD52L1
|
tumor protein D52-like 1 |
| chr4_-_15683118 | 0.12 |
ENST00000507899.1
ENST00000510802.1 |
FBXL5
|
F-box and leucine-rich repeat protein 5 |
| chr4_+_165675197 | 0.12 |
ENST00000515485.1
|
RP11-294O2.2
|
RP11-294O2.2 |
| chr11_+_827248 | 0.12 |
ENST00000527089.1
ENST00000530183.1 |
EFCAB4A
|
EF-hand calcium binding domain 4A |
| chr10_-_98347063 | 0.12 |
ENST00000443638.1
|
TM9SF3
|
transmembrane 9 superfamily member 3 |
| chr16_+_28996416 | 0.12 |
ENST00000395456.2
ENST00000454369.2 |
LAT
|
linker for activation of T cells |
| chr19_+_49977818 | 0.12 |
ENST00000594009.1
ENST00000595510.1 |
FLT3LG
|
fms-related tyrosine kinase 3 ligand |
| chr7_+_2281882 | 0.12 |
ENST00000397046.1
ENST00000397048.1 ENST00000454650.1 |
NUDT1
|
nudix (nucleoside diphosphate linked moiety X)-type motif 1 |
| chr2_+_120687335 | 0.12 |
ENST00000544261.1
|
PTPN4
|
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
| chr4_+_153701081 | 0.12 |
ENST00000451320.2
ENST00000429148.2 ENST00000353617.2 ENST00000405727.2 ENST00000356064.3 |
ARFIP1
|
ADP-ribosylation factor interacting protein 1 |
| chr22_-_37880543 | 0.12 |
ENST00000442496.1
|
MFNG
|
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr3_-_158390282 | 0.12 |
ENST00000264265.3
|
LXN
|
latexin |
| chr3_-_128879875 | 0.12 |
ENST00000418265.1
ENST00000393292.3 ENST00000273541.8 |
ISY1-RAB43
ISY1
|
ISY1-RAB43 readthrough ISY1 splicing factor homolog (S. cerevisiae) |
| chr1_+_59486059 | 0.11 |
ENST00000447329.1
|
RP4-794H19.4
|
RP4-794H19.4 |
| chr6_+_34433844 | 0.11 |
ENST00000244458.2
ENST00000374043.2 |
PACSIN1
|
protein kinase C and casein kinase substrate in neurons 1 |
| chr2_+_138721850 | 0.11 |
ENST00000329366.4
ENST00000280097.3 |
HNMT
|
histamine N-methyltransferase |
| chr6_+_125474939 | 0.11 |
ENST00000527711.1
|
TPD52L1
|
tumor protein D52-like 1 |
| chr6_-_130031358 | 0.11 |
ENST00000368149.2
|
ARHGAP18
|
Rho GTPase activating protein 18 |
| chr4_+_20702030 | 0.11 |
ENST00000510051.1
ENST00000503585.1 ENST00000360916.5 ENST00000295290.8 ENST00000514485.1 |
PACRGL
|
PARK2 co-regulated-like |
| chr1_+_212738676 | 0.11 |
ENST00000366981.4
ENST00000366987.2 |
ATF3
|
activating transcription factor 3 |
| chr5_+_892745 | 0.11 |
ENST00000166345.3
|
TRIP13
|
thyroid hormone receptor interactor 13 |
| chr11_-_104034827 | 0.11 |
ENST00000393158.2
|
PDGFD
|
platelet derived growth factor D |
| chr6_+_125475335 | 0.11 |
ENST00000532429.1
ENST00000534199.1 |
TPD52L1
|
tumor protein D52-like 1 |
| chr14_+_100531738 | 0.11 |
ENST00000555706.1
|
EVL
|
Enah/Vasp-like |
| chr6_-_31550192 | 0.11 |
ENST00000429299.2
ENST00000446745.2 |
LTB
|
lymphotoxin beta (TNF superfamily, member 3) |
| chr4_-_76598296 | 0.11 |
ENST00000395719.3
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr7_+_30589829 | 0.11 |
ENST00000579437.1
|
RP4-777O23.1
|
RP4-777O23.1 |
| chr3_-_47324242 | 0.11 |
ENST00000456548.1
ENST00000432493.1 ENST00000335044.2 ENST00000444589.2 |
KIF9
|
kinesin family member 9 |
| chr6_+_125474992 | 0.10 |
ENST00000528193.1
|
TPD52L1
|
tumor protein D52-like 1 |
| chr15_+_40453204 | 0.10 |
ENST00000287598.6
ENST00000412359.3 |
BUB1B
|
BUB1 mitotic checkpoint serine/threonine kinase B |
| chr17_-_8279977 | 0.10 |
ENST00000396267.1
|
KRBA2
|
KRAB-A domain containing 2 |
| chr15_+_71184931 | 0.10 |
ENST00000560369.1
ENST00000260382.5 |
LRRC49
|
leucine rich repeat containing 49 |
| chr19_+_41949054 | 0.10 |
ENST00000378187.2
|
C19orf69
|
chromosome 19 open reading frame 69 |
| chr6_-_29527702 | 0.10 |
ENST00000377050.4
|
UBD
|
ubiquitin D |
| chr11_+_47236489 | 0.10 |
ENST00000256996.4
ENST00000378603.3 ENST00000378600.3 ENST00000378601.3 |
DDB2
|
damage-specific DNA binding protein 2, 48kDa |
| chr17_-_56595196 | 0.10 |
ENST00000579921.1
ENST00000579925.1 ENST00000323456.5 |
MTMR4
|
myotubularin related protein 4 |
| chr17_-_62097927 | 0.10 |
ENST00000578313.1
ENST00000584084.1 ENST00000579788.1 ENST00000579687.1 ENST00000578379.1 ENST00000578892.1 ENST00000412356.1 ENST00000418105.1 |
ICAM2
|
intercellular adhesion molecule 2 |
| chr2_-_220041860 | 0.10 |
ENST00000453038.1
|
CNPPD1
|
cyclin Pas1/PHO80 domain containing 1 |
| chr1_-_161039753 | 0.10 |
ENST00000368015.1
|
ARHGAP30
|
Rho GTPase activating protein 30 |
| chr19_+_50431959 | 0.10 |
ENST00000595125.1
|
ATF5
|
activating transcription factor 5 |
| chr11_+_18154059 | 0.10 |
ENST00000531264.1
|
MRGPRX3
|
MAS-related GPR, member X3 |
| chr17_-_8263538 | 0.10 |
ENST00000535173.1
|
AC135178.1
|
HCG1985372; Uncharacterized protein; cDNA FLJ37541 fis, clone BRCAN2026340 |
| chr15_+_96875657 | 0.10 |
ENST00000559679.1
ENST00000394171.2 |
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr17_+_21729899 | 0.10 |
ENST00000583708.1
|
UBBP4
|
ubiquitin B pseudogene 4 |
| chr12_+_102513950 | 0.10 |
ENST00000378128.3
ENST00000327680.2 ENST00000541394.1 ENST00000543784.1 |
PARPBP
|
PARP1 binding protein |
| chr4_+_119199904 | 0.10 |
ENST00000602483.1
ENST00000602819.1 |
SNHG8
|
small nucleolar RNA host gene 8 (non-protein coding) |
| chr5_-_138862326 | 0.10 |
ENST00000330794.4
|
TMEM173
|
transmembrane protein 173 |
| chr16_+_28996114 | 0.10 |
ENST00000395461.3
|
LAT
|
linker for activation of T cells |
| chr16_-_57513657 | 0.10 |
ENST00000566936.1
ENST00000568617.1 ENST00000567276.1 ENST00000569548.1 ENST00000569250.1 ENST00000564378.1 |
DOK4
|
docking protein 4 |
| chr2_-_152118276 | 0.09 |
ENST00000409092.1
|
RBM43
|
RNA binding motif protein 43 |
| chr8_-_6115044 | 0.09 |
ENST00000519555.1
|
RP11-124B13.1
|
RP11-124B13.1 |
| chr22_-_29196546 | 0.09 |
ENST00000403532.3
ENST00000216037.6 |
XBP1
|
X-box binding protein 1 |
| chr8_-_125486755 | 0.09 |
ENST00000499418.2
ENST00000530778.1 |
RNF139-AS1
|
RNF139 antisense RNA 1 (head to head) |
| chr4_+_141264597 | 0.09 |
ENST00000338517.4
ENST00000394203.3 ENST00000506322.1 |
SCOC
|
short coiled-coil protein |
| chr3_-_121379739 | 0.09 |
ENST00000428394.2
ENST00000314583.3 |
HCLS1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr18_+_74240756 | 0.09 |
ENST00000584910.1
ENST00000582452.1 |
LINC00908
|
long intergenic non-protein coding RNA 908 |
| chr19_-_39368887 | 0.09 |
ENST00000340740.3
ENST00000591812.1 |
RINL
|
Ras and Rab interactor-like |
| chr20_-_52612705 | 0.09 |
ENST00000434986.2
|
BCAS1
|
breast carcinoma amplified sequence 1 |
| chr17_+_21729593 | 0.09 |
ENST00000581769.1
ENST00000584755.1 |
UBBP4
|
ubiquitin B pseudogene 4 |
| chr18_-_72265035 | 0.09 |
ENST00000585279.1
ENST00000580048.1 |
LINC00909
|
long intergenic non-protein coding RNA 909 |
| chr8_+_143761874 | 0.09 |
ENST00000301258.4
ENST00000513264.1 |
PSCA
|
prostate stem cell antigen |
| chr17_-_4689649 | 0.09 |
ENST00000441199.2
ENST00000416307.2 |
VMO1
|
vitelline membrane outer layer 1 homolog (chicken) |
| chr9_-_104249400 | 0.09 |
ENST00000374848.3
|
TMEM246
|
transmembrane protein 246 |
| chr7_-_944631 | 0.09 |
ENST00000453175.2
|
ADAP1
|
ArfGAP with dual PH domains 1 |
| chr10_+_112631699 | 0.09 |
ENST00000444997.1
|
PDCD4
|
programmed cell death 4 (neoplastic transformation inhibitor) |
| chr10_-_75255724 | 0.09 |
ENST00000342558.3
ENST00000360663.5 ENST00000394829.2 ENST00000394828.2 ENST00000394822.2 |
PPP3CB
|
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr19_-_42192189 | 0.09 |
ENST00000401731.1
ENST00000338196.4 ENST00000006724.3 |
CEACAM7
|
carcinoembryonic antigen-related cell adhesion molecule 7 |
| chr3_-_179169181 | 0.09 |
ENST00000497513.1
|
GNB4
|
guanine nucleotide binding protein (G protein), beta polypeptide 4 |
| chr16_-_3149278 | 0.09 |
ENST00000575108.1
ENST00000576483.1 ENST00000538082.2 ENST00000576985.1 |
ZSCAN10
|
zinc finger and SCAN domain containing 10 |
| chr2_+_99771527 | 0.09 |
ENST00000415142.1
ENST00000436234.1 |
LIPT1
|
lipoyltransferase 1 |
| chr1_+_2477831 | 0.09 |
ENST00000606645.1
|
RP3-395M20.12
|
RP3-395M20.12 |
| chr9_-_139268068 | 0.09 |
ENST00000371734.3
ENST00000371732.5 ENST00000315908.7 |
CARD9
|
caspase recruitment domain family, member 9 |
| chr15_-_90234006 | 0.09 |
ENST00000300056.3
ENST00000559170.1 |
PEX11A
|
peroxisomal biogenesis factor 11 alpha |
| chr15_+_71185148 | 0.09 |
ENST00000443425.2
ENST00000560755.1 |
LRRC49
|
leucine rich repeat containing 49 |
| chr2_-_175499294 | 0.08 |
ENST00000392547.2
|
WIPF1
|
WAS/WASL interacting protein family, member 1 |
| chr7_-_6746474 | 0.08 |
ENST00000394917.3
ENST00000405858.1 ENST00000342651.5 |
ZNF12
|
zinc finger protein 12 |
| chr6_-_31745037 | 0.08 |
ENST00000375688.4
|
VWA7
|
von Willebrand factor A domain containing 7 |
| chr5_+_61874562 | 0.08 |
ENST00000334994.5
ENST00000409534.1 |
LRRC70
IPO11
|
leucine rich repeat containing 70 importin 11 |
| chr11_-_104035088 | 0.08 |
ENST00000302251.5
|
PDGFD
|
platelet derived growth factor D |
| chr5_-_130970723 | 0.08 |
ENST00000308008.6
ENST00000296859.6 ENST00000507093.1 ENST00000510071.1 ENST00000509018.1 ENST00000307984.5 |
RAPGEF6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr11_+_18344106 | 0.08 |
ENST00000534641.1
ENST00000525831.1 ENST00000265963.4 |
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa |
| chr6_+_27925019 | 0.08 |
ENST00000244623.1
|
OR2B6
|
olfactory receptor, family 2, subfamily B, member 6 |
| chr4_-_140477928 | 0.08 |
ENST00000274031.3
|
SETD7
|
SET domain containing (lysine methyltransferase) 7 |
| chr2_-_242556900 | 0.08 |
ENST00000402545.1
ENST00000402136.1 |
THAP4
|
THAP domain containing 4 |
| chr14_-_45722605 | 0.08 |
ENST00000310806.4
|
MIS18BP1
|
MIS18 binding protein 1 |
| chr8_-_82598067 | 0.08 |
ENST00000523942.1
ENST00000522997.1 |
IMPA1
|
inositol(myo)-1(or 4)-monophosphatase 1 |
| chr12_-_49523896 | 0.08 |
ENST00000549870.1
|
TUBA1B
|
tubulin, alpha 1b |
| chr8_-_42396721 | 0.08 |
ENST00000518384.1
|
SLC20A2
|
solute carrier family 20 (phosphate transporter), member 2 |
| chr11_-_46722117 | 0.08 |
ENST00000311956.4
|
ARHGAP1
|
Rho GTPase activating protein 1 |
| chr6_-_34855773 | 0.08 |
ENST00000420584.2
ENST00000361288.4 |
TAF11
|
TAF11 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 28kDa |
| chr1_+_10509971 | 0.08 |
ENST00000320498.4
|
CORT
|
cortistatin |
| chr22_-_29196030 | 0.08 |
ENST00000405219.3
|
XBP1
|
X-box binding protein 1 |
| chr9_-_77567743 | 0.08 |
ENST00000376854.5
|
C9orf40
|
chromosome 9 open reading frame 40 |
| chr3_+_113465866 | 0.08 |
ENST00000273398.3
ENST00000538620.1 ENST00000496747.1 ENST00000475322.1 |
ATP6V1A
|
ATPase, H+ transporting, lysosomal 70kDa, V1 subunit A |
| chr7_+_33765593 | 0.08 |
ENST00000311067.3
|
RP11-89N17.1
|
HCG1643653; Uncharacterized protein |
| chr16_+_21312170 | 0.08 |
ENST00000338573.5
ENST00000561968.1 |
CRYM-AS1
|
CRYM antisense RNA 1 |
| chr6_+_10528560 | 0.08 |
ENST00000379597.3
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr19_+_41281282 | 0.08 |
ENST00000263369.3
|
MIA
|
melanoma inhibitory activity |
| chr12_-_133405288 | 0.08 |
ENST00000204726.3
|
GOLGA3
|
golgin A3 |
| chr16_+_20775358 | 0.08 |
ENST00000440284.2
|
ACSM3
|
acyl-CoA synthetase medium-chain family member 3 |
| chr19_+_36393367 | 0.08 |
ENST00000246551.4
|
HCST
|
hematopoietic cell signal transducer |
| chr5_+_140739537 | 0.08 |
ENST00000522605.1
|
PCDHGB2
|
protocadherin gamma subfamily B, 2 |
| chr19_+_58111241 | 0.08 |
ENST00000597700.1
ENST00000332854.6 ENST00000597864.1 |
ZNF530
|
zinc finger protein 530 |
| chr7_+_79763271 | 0.08 |
ENST00000442586.1
|
GNAI1
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
| chr11_+_61976137 | 0.08 |
ENST00000244930.4
|
SCGB2A1
|
secretoglobin, family 2A, member 1 |
| chr3_-_20227619 | 0.08 |
ENST00000425061.1
ENST00000443724.1 ENST00000421451.1 ENST00000452020.1 ENST00000417364.1 ENST00000306698.2 ENST00000419233.2 ENST00000263753.4 ENST00000383774.1 ENST00000437051.1 ENST00000412868.1 ENST00000429446.3 ENST00000442720.1 |
SGOL1
|
shugoshin-like 1 (S. pombe) |
| chr1_+_234509413 | 0.08 |
ENST00000366613.1
ENST00000366612.1 |
COA6
|
cytochrome c oxidase assembly factor 6 homolog (S. cerevisiae) |
| chrX_-_131352152 | 0.08 |
ENST00000342983.2
|
RAP2C
|
RAP2C, member of RAS oncogene family |
| chr18_-_12377001 | 0.07 |
ENST00000590811.1
|
AFG3L2
|
AFG3-like AAA ATPase 2 |
| chr20_+_34802295 | 0.07 |
ENST00000432603.1
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr1_+_63989004 | 0.07 |
ENST00000371088.4
|
EFCAB7
|
EF-hand calcium binding domain 7 |
| chr2_+_3642545 | 0.07 |
ENST00000382062.2
ENST00000236693.7 ENST00000349077.4 |
COLEC11
|
collectin sub-family member 11 |
| chrX_+_54556633 | 0.07 |
ENST00000336470.4
ENST00000360845.2 |
GNL3L
|
guanine nucleotide binding protein-like 3 (nucleolar)-like |
| chr11_-_28129656 | 0.07 |
ENST00000263181.6
|
KIF18A
|
kinesin family member 18A |
| chr16_-_30393752 | 0.07 |
ENST00000566517.1
ENST00000605106.1 |
SEPT1
SEPT1
|
septin 1 Uncharacterized protein |
| chr16_+_66442411 | 0.07 |
ENST00000499966.1
|
LINC00920
|
long intergenic non-protein coding RNA 920 |
| chr12_+_72058130 | 0.07 |
ENST00000547843.1
|
THAP2
|
THAP domain containing, apoptosis associated protein 2 |
| chr20_+_43104508 | 0.07 |
ENST00000262605.4
ENST00000372904.3 |
TTPAL
|
tocopherol (alpha) transfer protein-like |
| chr7_+_134212312 | 0.07 |
ENST00000359579.4
|
AKR1B10
|
aldo-keto reductase family 1, member B10 (aldose reductase) |
| chr4_-_84205905 | 0.07 |
ENST00000311461.7
ENST00000311469.4 ENST00000439031.2 |
COQ2
|
coenzyme Q2 4-hydroxybenzoate polyprenyltransferase |
| chr5_-_43515125 | 0.07 |
ENST00000509489.1
|
C5orf34
|
chromosome 5 open reading frame 34 |
| chr14_-_107083690 | 0.07 |
ENST00000455737.1
ENST00000390629.2 |
IGHV4-59
|
immunoglobulin heavy variable 4-59 |
| chr11_-_72432950 | 0.07 |
ENST00000426523.1
ENST00000429686.1 |
ARAP1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr15_-_90233866 | 0.07 |
ENST00000561257.1
|
PEX11A
|
peroxisomal biogenesis factor 11 alpha |
| chr12_-_15374343 | 0.07 |
ENST00000256953.2
ENST00000546331.1 |
RERG
|
RAS-like, estrogen-regulated, growth inhibitor |
| chr4_+_2800722 | 0.07 |
ENST00000508385.1
|
SH3BP2
|
SH3-domain binding protein 2 |
| chr12_+_6602517 | 0.07 |
ENST00000315579.5
ENST00000539714.1 |
NCAPD2
|
non-SMC condensin I complex, subunit D2 |
| chr1_+_24285599 | 0.07 |
ENST00000471915.1
|
PNRC2
|
proline-rich nuclear receptor coactivator 2 |
| chr3_+_44690211 | 0.07 |
ENST00000396056.2
ENST00000432115.2 ENST00000415571.2 ENST00000399560.2 ENST00000296092.3 ENST00000542250.1 ENST00000453164.1 |
ZNF35
|
zinc finger protein 35 |
| chr3_+_118892362 | 0.07 |
ENST00000497685.1
ENST00000264234.3 |
UPK1B
|
uroplakin 1B |
| chr1_-_55230165 | 0.07 |
ENST00000371279.3
|
PARS2
|
prolyl-tRNA synthetase 2, mitochondrial (putative) |
| chr19_+_41281060 | 0.07 |
ENST00000594436.1
ENST00000597784.1 |
MIA
|
melanoma inhibitory activity |
| chr1_+_28099700 | 0.07 |
ENST00000440806.2
|
STX12
|
syntaxin 12 |
| chr17_+_7211656 | 0.07 |
ENST00000416016.2
|
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr11_-_64013288 | 0.07 |
ENST00000542235.1
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr1_+_6086380 | 0.07 |
ENST00000602612.1
ENST00000378087.3 ENST00000341524.1 ENST00000352527.1 |
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
| chr12_-_105629852 | 0.07 |
ENST00000551662.1
ENST00000553097.1 |
APPL2
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
| chr1_+_225965518 | 0.07 |
ENST00000304786.7
ENST00000366839.4 ENST00000366838.1 |
SRP9
|
signal recognition particle 9kDa |
| chr2_+_217082311 | 0.07 |
ENST00000597904.1
|
RP11-566E18.3
|
RP11-566E18.3 |
| chr5_+_172386419 | 0.07 |
ENST00000265100.2
ENST00000519239.1 |
RPL26L1
|
ribosomal protein L26-like 1 |
| chr12_+_49297899 | 0.07 |
ENST00000552942.1
ENST00000320516.4 |
CCDC65
|
coiled-coil domain containing 65 |
| chr5_+_102455968 | 0.07 |
ENST00000358359.3
|
PPIP5K2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr11_+_86667117 | 0.07 |
ENST00000531827.1
|
RP11-736K20.6
|
RP11-736K20.6 |
| chr20_+_43104541 | 0.07 |
ENST00000372906.2
ENST00000456317.1 |
TTPAL
|
tocopherol (alpha) transfer protein-like |
| chr5_+_39520499 | 0.07 |
ENST00000604954.1
|
CTD-2078B5.2
|
CTD-2078B5.2 |
| chr20_+_30327063 | 0.07 |
ENST00000300403.6
ENST00000340513.4 |
TPX2
|
TPX2, microtubule-associated |
| chr1_+_207277632 | 0.07 |
ENST00000421786.1
|
C4BPA
|
complement component 4 binding protein, alpha |
| chr6_+_52535878 | 0.07 |
ENST00000211314.4
|
TMEM14A
|
transmembrane protein 14A |
| chr2_-_220041617 | 0.07 |
ENST00000451647.1
ENST00000360507.5 |
CNPPD1
|
cyclin Pas1/PHO80 domain containing 1 |
| chr6_-_31745085 | 0.07 |
ENST00000375686.3
ENST00000447450.1 |
VWA7
|
von Willebrand factor A domain containing 7 |
| chr1_-_151254362 | 0.07 |
ENST00000447795.2
|
RP11-126K1.2
|
Uncharacterized protein |
| chr22_+_21311367 | 0.07 |
ENST00000444039.1
|
XXbac-B135H6.15
|
XXbac-B135H6.15 |
| chr6_-_119031228 | 0.07 |
ENST00000392500.3
ENST00000368488.5 ENST00000434604.1 |
CEP85L
|
centrosomal protein 85kDa-like |
| chr2_+_219472637 | 0.07 |
ENST00000417849.1
|
PLCD4
|
phospholipase C, delta 4 |
| chr17_+_4618734 | 0.07 |
ENST00000571206.1
|
ARRB2
|
arrestin, beta 2 |
| chr1_+_84767289 | 0.07 |
ENST00000394834.3
ENST00000370669.1 |
SAMD13
|
sterile alpha motif domain containing 13 |
| chr4_+_128802016 | 0.07 |
ENST00000270861.5
ENST00000515069.1 ENST00000513090.1 ENST00000507249.1 |
PLK4
|
polo-like kinase 4 |
| chr7_-_37382683 | 0.07 |
ENST00000455879.1
|
ELMO1
|
engulfment and cell motility 1 |
| chr7_+_37960163 | 0.07 |
ENST00000199448.4
ENST00000559325.1 ENST00000423717.1 |
EPDR1
|
ependymin related 1 |
| chr7_-_32529973 | 0.07 |
ENST00000410044.1
ENST00000409987.1 ENST00000409782.1 ENST00000450169.2 |
LSM5
|
LSM5 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr1_-_101360374 | 0.07 |
ENST00000535414.1
|
EXTL2
|
exostosin-like glycosyltransferase 2 |
| chr11_-_71791726 | 0.07 |
ENST00000393695.3
|
NUMA1
|
nuclear mitotic apparatus protein 1 |
| chr15_+_82555125 | 0.07 |
ENST00000566205.1
ENST00000339465.5 ENST00000569120.1 ENST00000566861.1 |
FAM154B
|
family with sequence similarity 154, member B |
| chr9_-_104249319 | 0.07 |
ENST00000374847.1
|
TMEM246
|
transmembrane protein 246 |
| chr5_-_175388120 | 0.07 |
ENST00000515016.1
|
THOC3
|
THO complex 3 |
| chr9_-_33264557 | 0.07 |
ENST00000473781.1
ENST00000488499.1 |
BAG1
|
BCL2-associated athanogene |
| chr6_-_108278456 | 0.07 |
ENST00000429168.1
|
SEC63
|
SEC63 homolog (S. cerevisiae) |
| chr22_-_29196511 | 0.07 |
ENST00000344347.5
|
XBP1
|
X-box binding protein 1 |
| chr4_+_119199864 | 0.06 |
ENST00000602414.1
ENST00000602520.1 |
SNHG8
|
small nucleolar RNA host gene 8 (non-protein coding) |
| chr8_-_144660771 | 0.06 |
ENST00000449291.2
|
NAPRT1
|
nicotinate phosphoribosyltransferase domain containing 1 |
| chr7_-_72971934 | 0.06 |
ENST00000411832.1
|
BCL7B
|
B-cell CLL/lymphoma 7B |
| chr1_+_45212051 | 0.06 |
ENST00000372222.3
|
KIF2C
|
kinesin family member 2C |
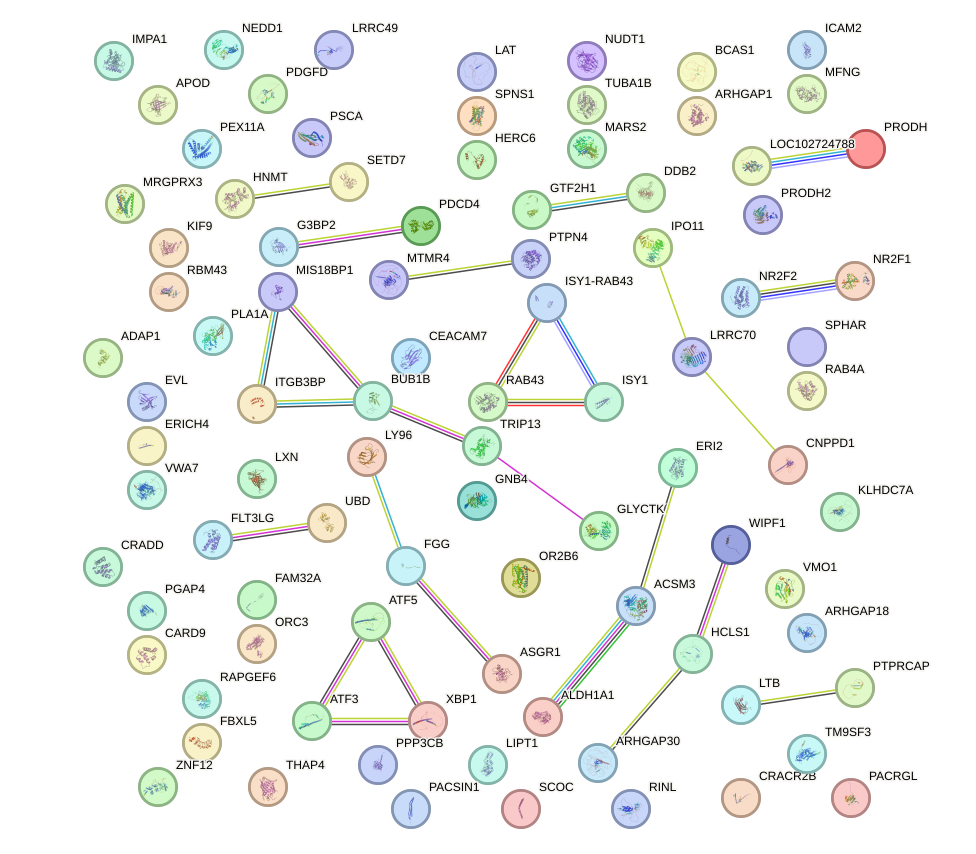
Network of associatons between targets according to the STRING database.
First level regulatory network of FLI1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1903489 | epithelial cell differentiation involved in salivary gland development(GO:0060690) epithelial cell maturation involved in salivary gland development(GO:0060691) regulation of plasma cell differentiation(GO:1900098) positive regulation of plasma cell differentiation(GO:1900100) regulation of lactation(GO:1903487) positive regulation of lactation(GO:1903489) |
| 0.1 | 0.2 | GO:0035696 | monocyte extravasation(GO:0035696) regulation of monocyte extravasation(GO:2000437) |
| 0.1 | 0.3 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.1 | GO:1904761 | negative regulation of myofibroblast differentiation(GO:1904761) |
| 0.0 | 0.2 | GO:0006203 | dGTP catabolic process(GO:0006203) |
| 0.0 | 0.2 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.2 | GO:0000350 | generation of catalytic spliceosome for second transesterification step(GO:0000350) |
| 0.0 | 0.2 | GO:0010133 | proline catabolic process(GO:0006562) proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.1 | GO:0034443 | negative regulation of lipoprotein oxidation(GO:0034443) |
| 0.0 | 0.1 | GO:0030885 | regulation of myeloid dendritic cell activation(GO:0030885) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:0006427 | histidyl-tRNA aminoacylation(GO:0006427) |
| 0.0 | 0.1 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.1 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 0.1 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.1 | GO:0016107 | sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) |
| 0.0 | 0.1 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.0 | 0.1 | GO:0006425 | glutaminyl-tRNA aminoacylation(GO:0006425) |
| 0.0 | 0.1 | GO:0006433 | prolyl-tRNA aminoacylation(GO:0006433) |
| 0.0 | 0.1 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 0.0 | 0.0 | GO:1990564 | protein ufmylation(GO:0071569) protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.1 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.0 | 0.1 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.0 | 0.1 | GO:0070898 | RNA polymerase III transcriptional preinitiation complex assembly(GO:0070898) |
| 0.0 | 0.1 | GO:0002590 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) |
| 0.0 | 0.1 | GO:0003273 | cell migration involved in endocardial cushion formation(GO:0003273) |
| 0.0 | 0.1 | GO:0030037 | actin filament reorganization involved in cell cycle(GO:0030037) |
| 0.0 | 0.1 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.0 | 0.1 | GO:0002904 | B cell negative selection(GO:0002352) positive regulation of B cell apoptotic process(GO:0002904) post-embryonic camera-type eye morphogenesis(GO:0048597) apoptotic process involved in embryonic digit morphogenesis(GO:1902263) positive regulation of apoptotic DNA fragmentation(GO:1902512) |
| 0.0 | 0.1 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.0 | 0.1 | GO:0044837 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.1 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.0 | 0.1 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.0 | 0.1 | GO:0006742 | NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.0 | 0.2 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.0 | 0.1 | GO:0044375 | peroxisome membrane biogenesis(GO:0016557) regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.2 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.1 | GO:0035669 | TRAM-dependent toll-like receptor signaling pathway(GO:0035668) TRAM-dependent toll-like receptor 4 signaling pathway(GO:0035669) |
| 0.0 | 0.2 | GO:0032888 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.0 | 0.1 | GO:1902299 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.5 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.0 | 0.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.2 | GO:0010032 | meiotic chromosome condensation(GO:0010032) |
| 0.0 | 0.1 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.1 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.0 | 0.0 | GO:0046041 | ITP metabolic process(GO:0046041) |
| 0.0 | 0.2 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.1 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.0 | 0.0 | GO:0034756 | regulation of iron ion transport(GO:0034756) regulation of iron ion transmembrane transport(GO:0034759) |
| 0.0 | 0.0 | GO:0061110 | dense core granule biogenesis(GO:0061110) regulation of dense core granule biogenesis(GO:2000705) |
| 0.0 | 0.0 | GO:0010728 | regulation of hydrogen peroxide biosynthetic process(GO:0010728) |
| 0.0 | 0.0 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) gall bladder development(GO:0061010) |
| 0.0 | 0.1 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.0 | 0.0 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.0 | 0.0 | GO:0048075 | positive regulation of eye pigmentation(GO:0048075) |
| 0.0 | 0.0 | GO:0045210 | negative regulation of dendritic cell cytokine production(GO:0002731) FasL biosynthetic process(GO:0045210) |
| 0.0 | 0.0 | GO:0061582 | intestinal epithelial cell migration(GO:0061582) |
| 0.0 | 0.1 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 | 0.0 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) siRNA loading onto RISC involved in RNA interference(GO:0035087) |
| 0.0 | 0.0 | GO:0043000 | Golgi to plasma membrane CFTR protein transport(GO:0043000) |
| 0.0 | 0.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.1 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0071020 | post-spliceosomal complex(GO:0071020) |
| 0.0 | 0.1 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.1 | GO:0072563 | endothelial microparticle(GO:0072563) |
| 0.0 | 0.1 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.1 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.1 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.0 | 0.1 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.0 | 0.2 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.0 | 0.1 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.0 | 0.1 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.0 | 0.1 | GO:0031379 | RNA-directed RNA polymerase complex(GO:0031379) |
| 0.0 | 0.0 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.2 | GO:0055028 | cortical microtubule(GO:0055028) |
| 0.0 | 0.1 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.2 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.1 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.2 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.4 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.1 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.0 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.0 | 0.0 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0008413 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 0.1 | 0.5 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) |
| 0.1 | 0.2 | GO:0000384 | first spliceosomal transesterification activity(GO:0000384) |
| 0.1 | 0.2 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 0.2 | GO:0035248 | alpha-1,4-N-acetylgalactosaminyltransferase activity(GO:0035248) |
| 0.0 | 0.3 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.0 | 0.1 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 0.0 | 0.2 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.2 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.2 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.0 | 0.1 | GO:0045550 | geranylgeranyl reductase activity(GO:0045550) |
| 0.0 | 0.1 | GO:0000992 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) |
| 0.0 | 0.1 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0004827 | proline-tRNA ligase activity(GO:0004827) |
| 0.0 | 0.1 | GO:0031859 | platelet activating factor receptor binding(GO:0031859) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.1 | GO:0034057 | RNA strand-exchange activity(GO:0034057) |
| 0.0 | 0.1 | GO:0016979 | lipoate-protein ligase activity(GO:0016979) |
| 0.0 | 0.1 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.2 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.0 | GO:0031691 | alpha-1A adrenergic receptor binding(GO:0031691) follicle-stimulating hormone receptor binding(GO:0031762) V2 vasopressin receptor binding(GO:0031896) |
| 0.0 | 0.0 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.0 | 0.0 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.3 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.0 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.0 | GO:0017130 | poly(C) RNA binding(GO:0017130) |
| 0.0 | 0.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.1 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 0.1 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.0 | 0.0 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.1 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.1 | GO:0031403 | lithium ion binding(GO:0031403) |
| 0.0 | 0.1 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.2 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.3 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.5 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.1 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |