Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
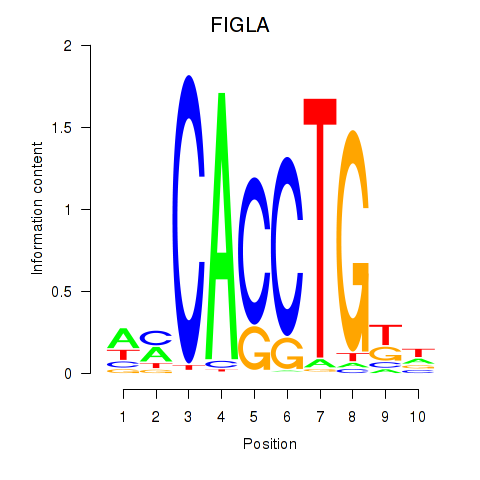
Results for FIGLA
Z-value: 0.73
Transcription factors associated with FIGLA
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
FIGLA
|
ENSG00000183733.6 | folliculogenesis specific bHLH transcription factor |
Activity profile of FIGLA motif
Sorted Z-values of FIGLA motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_48133459 | 1.03 |
ENST00000320031.8
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) |
| chr2_-_42160486 | 0.95 |
ENST00000427054.1
|
AC104654.2
|
AC104654.2 |
| chr15_+_45722727 | 0.92 |
ENST00000396650.2
ENST00000558435.1 ENST00000344300.3 |
C15orf48
|
chromosome 15 open reading frame 48 |
| chr17_+_48133330 | 0.90 |
ENST00000544892.1
|
ITGA3
|
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) |
| chr3_-_49851313 | 0.81 |
ENST00000333486.3
|
UBA7
|
ubiquitin-like modifier activating enzyme 7 |
| chr2_+_121493717 | 0.78 |
ENST00000418323.1
|
GLI2
|
GLI family zinc finger 2 |
| chr1_+_183155373 | 0.63 |
ENST00000493293.1
ENST00000264144.4 |
LAMC2
|
laminin, gamma 2 |
| chr5_+_34656529 | 0.60 |
ENST00000513974.1
ENST00000512629.1 |
RAI14
|
retinoic acid induced 14 |
| chr14_-_37051798 | 0.60 |
ENST00000258829.5
|
NKX2-8
|
NK2 homeobox 8 |
| chr22_-_50970506 | 0.58 |
ENST00000428989.2
ENST00000403326.1 |
ODF3B
|
outer dense fiber of sperm tails 3B |
| chr15_+_89182156 | 0.53 |
ENST00000379224.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr5_+_34656569 | 0.52 |
ENST00000428746.2
|
RAI14
|
retinoic acid induced 14 |
| chr15_+_89182178 | 0.52 |
ENST00000559876.1
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr11_-_33913708 | 0.52 |
ENST00000257818.2
|
LMO2
|
LIM domain only 2 (rhombotin-like 1) |
| chr11_+_69455855 | 0.43 |
ENST00000227507.2
ENST00000536559.1 |
CCND1
|
cyclin D1 |
| chr15_+_89181974 | 0.42 |
ENST00000306072.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr1_-_153585539 | 0.39 |
ENST00000368706.4
|
S100A16
|
S100 calcium binding protein A16 |
| chr2_+_101437487 | 0.39 |
ENST00000427413.1
ENST00000542504.1 |
NPAS2
|
neuronal PAS domain protein 2 |
| chr5_-_141257954 | 0.37 |
ENST00000456271.1
ENST00000394536.3 ENST00000503492.1 ENST00000287008.3 |
PCDH1
|
protocadherin 1 |
| chr1_+_110754094 | 0.36 |
ENST00000369787.3
ENST00000413138.3 ENST00000438661.2 |
KCNC4
|
potassium voltage-gated channel, Shaw-related subfamily, member 4 |
| chr17_+_68165657 | 0.36 |
ENST00000243457.3
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr8_-_123706338 | 0.35 |
ENST00000521608.1
|
RP11-973F15.1
|
long intergenic non-protein coding RNA 1151 |
| chr11_-_124632179 | 0.34 |
ENST00000278927.5
ENST00000442070.2 ENST00000444566.1 ENST00000435477.1 |
ESAM
|
endothelial cell adhesion molecule |
| chr5_-_172755056 | 0.33 |
ENST00000520648.1
|
STC2
|
stanniocalcin 2 |
| chr7_+_12726623 | 0.32 |
ENST00000439721.1
|
ARL4A
|
ADP-ribosylation factor-like 4A |
| chr10_-_69991865 | 0.31 |
ENST00000373673.3
|
ATOH7
|
atonal homolog 7 (Drosophila) |
| chr18_+_77439775 | 0.31 |
ENST00000299543.7
ENST00000075430.7 |
CTDP1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1 |
| chr15_+_90728145 | 0.30 |
ENST00000561085.1
ENST00000379122.3 ENST00000332496.6 |
SEMA4B
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr1_-_150669604 | 0.30 |
ENST00000427665.1
ENST00000540514.1 |
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr16_+_30006997 | 0.29 |
ENST00000304516.7
|
INO80E
|
INO80 complex subunit E |
| chr14_+_23025534 | 0.29 |
ENST00000557595.1
|
AE000662.92
|
Uncharacterized protein |
| chr16_+_30006615 | 0.28 |
ENST00000563197.1
|
INO80E
|
INO80 complex subunit E |
| chr6_-_36355513 | 0.28 |
ENST00000340181.4
ENST00000373737.4 |
ETV7
|
ets variant 7 |
| chr6_-_133055896 | 0.28 |
ENST00000367927.5
ENST00000425515.2 ENST00000207771.3 ENST00000392393.3 ENST00000450865.2 ENST00000392394.2 |
VNN3
|
vanin 3 |
| chr6_-_19804973 | 0.27 |
ENST00000457670.1
ENST00000607810.1 ENST00000606628.1 |
RP4-625H18.2
|
RP4-625H18.2 |
| chr3_-_48594248 | 0.27 |
ENST00000545984.1
ENST00000232375.3 ENST00000416568.1 ENST00000383734.2 ENST00000541519.1 ENST00000412035.1 |
PFKFB4
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 |
| chr11_-_129872672 | 0.25 |
ENST00000531431.1
ENST00000527581.1 |
PRDM10
|
PR domain containing 10 |
| chr4_-_140223670 | 0.25 |
ENST00000394228.1
ENST00000539387.1 |
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr17_+_40985407 | 0.25 |
ENST00000586114.1
ENST00000590720.1 ENST00000585805.1 ENST00000541124.1 ENST00000441946.2 ENST00000591152.1 ENST00000589469.1 ENST00000293362.3 ENST00000592169.1 |
PSME3
|
proteasome (prosome, macropain) activator subunit 3 (PA28 gamma; Ki) |
| chr5_+_34656331 | 0.24 |
ENST00000265109.3
|
RAI14
|
retinoic acid induced 14 |
| chr19_+_35940486 | 0.24 |
ENST00000246549.2
|
FFAR2
|
free fatty acid receptor 2 |
| chr16_-_103572 | 0.24 |
ENST00000293860.5
|
POLR3K
|
polymerase (RNA) III (DNA directed) polypeptide K, 12.3 kDa |
| chr4_-_140223614 | 0.24 |
ENST00000394223.1
|
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr1_-_156675368 | 0.23 |
ENST00000368222.3
|
CRABP2
|
cellular retinoic acid binding protein 2 |
| chr12_+_122150646 | 0.23 |
ENST00000449592.2
|
TMEM120B
|
transmembrane protein 120B |
| chr22_-_42310570 | 0.23 |
ENST00000457093.1
|
SHISA8
|
shisa family member 8 |
| chr5_-_150467221 | 0.23 |
ENST00000522226.1
|
TNIP1
|
TNFAIP3 interacting protein 1 |
| chrX_+_53078465 | 0.23 |
ENST00000375466.2
|
GPR173
|
G protein-coupled receptor 173 |
| chr5_-_150466692 | 0.22 |
ENST00000315050.7
ENST00000523338.1 ENST00000522100.1 |
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr19_+_36347787 | 0.22 |
ENST00000347900.6
ENST00000360202.5 |
KIRREL2
|
kin of IRRE like 2 (Drosophila) |
| chr6_-_133055815 | 0.22 |
ENST00000509351.1
ENST00000417437.2 ENST00000414302.2 ENST00000423615.2 ENST00000427187.2 ENST00000275223.3 ENST00000519686.2 |
VNN3
|
vanin 3 |
| chr10_+_94608218 | 0.22 |
ENST00000371543.1
|
EXOC6
|
exocyst complex component 6 |
| chr17_-_48133054 | 0.22 |
ENST00000499842.1
|
RP11-1094H24.4
|
RP11-1094H24.4 |
| chr2_+_220299547 | 0.22 |
ENST00000312358.7
|
SPEG
|
SPEG complex locus |
| chr17_-_39183452 | 0.22 |
ENST00000361883.5
|
KRTAP1-5
|
keratin associated protein 1-5 |
| chr16_-_69418553 | 0.21 |
ENST00000569542.2
|
TERF2
|
telomeric repeat binding factor 2 |
| chr5_+_34656450 | 0.21 |
ENST00000514527.1
|
RAI14
|
retinoic acid induced 14 |
| chr6_-_19804877 | 0.21 |
ENST00000447250.1
|
RP4-625H18.2
|
RP4-625H18.2 |
| chr2_-_75788038 | 0.21 |
ENST00000393913.3
ENST00000410113.1 |
EVA1A
|
eva-1 homolog A (C. elegans) |
| chr1_-_156675564 | 0.21 |
ENST00000368220.1
|
CRABP2
|
cellular retinoic acid binding protein 2 |
| chr17_-_39211463 | 0.21 |
ENST00000542910.1
ENST00000398477.1 |
KRTAP2-2
|
keratin associated protein 2-2 |
| chr7_+_35756186 | 0.20 |
ENST00000430518.1
|
AC018647.3
|
AC018647.3 |
| chr1_-_11918988 | 0.20 |
ENST00000376468.3
|
NPPB
|
natriuretic peptide B |
| chr2_+_177001685 | 0.20 |
ENST00000432796.2
|
HOXD3
|
homeobox D3 |
| chr7_+_130794878 | 0.20 |
ENST00000416992.2
|
MKLN1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr19_-_46272106 | 0.20 |
ENST00000560168.1
|
SIX5
|
SIX homeobox 5 |
| chr17_-_39216344 | 0.20 |
ENST00000391418.2
|
KRTAP2-3
|
keratin associated protein 2-3 |
| chr19_+_54385439 | 0.20 |
ENST00000536044.1
ENST00000540413.1 ENST00000263431.3 ENST00000419486.1 |
PRKCG
|
protein kinase C, gamma |
| chr17_-_39203519 | 0.19 |
ENST00000542137.1
ENST00000391419.3 |
KRTAP2-1
|
keratin associated protein 2-1 |
| chr5_-_94620239 | 0.19 |
ENST00000515393.1
|
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr2_-_154335300 | 0.19 |
ENST00000325926.3
|
RPRM
|
reprimo, TP53 dependent G2 arrest mediator candidate |
| chr19_+_55996565 | 0.18 |
ENST00000587400.1
|
NAT14
|
N-acetyltransferase 14 (GCN5-related, putative) |
| chr6_-_36355486 | 0.18 |
ENST00000538992.1
|
ETV7
|
ets variant 7 |
| chr22_+_31090793 | 0.18 |
ENST00000332585.6
ENST00000382310.3 ENST00000446658.2 |
OSBP2
|
oxysterol binding protein 2 |
| chr6_+_158402860 | 0.18 |
ENST00000367122.2
ENST00000367121.3 ENST00000355585.4 ENST00000367113.4 |
SYNJ2
|
synaptojanin 2 |
| chr7_-_8301869 | 0.18 |
ENST00000402384.3
|
ICA1
|
islet cell autoantigen 1, 69kDa |
| chr15_+_91411810 | 0.18 |
ENST00000268171.3
|
FURIN
|
furin (paired basic amino acid cleaving enzyme) |
| chr3_-_194991876 | 0.18 |
ENST00000310380.6
|
XXYLT1
|
xyloside xylosyltransferase 1 |
| chr6_+_31783291 | 0.18 |
ENST00000375651.5
ENST00000608703.1 ENST00000458062.2 |
HSPA1A
|
heat shock 70kDa protein 1A |
| chr17_+_79495397 | 0.17 |
ENST00000417245.2
ENST00000334850.7 |
FSCN2
|
fascin homolog 2, actin-bundling protein, retinal (Strongylocentrotus purpuratus) |
| chr11_-_119252359 | 0.17 |
ENST00000455332.2
|
USP2
|
ubiquitin specific peptidase 2 |
| chr8_+_32405785 | 0.17 |
ENST00000287842.3
|
NRG1
|
neuregulin 1 |
| chr3_+_49059038 | 0.17 |
ENST00000451378.2
|
NDUFAF3
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 3 |
| chr6_+_31939608 | 0.17 |
ENST00000375331.2
ENST00000375333.2 |
STK19
|
serine/threonine kinase 19 |
| chr8_-_49833978 | 0.17 |
ENST00000020945.1
|
SNAI2
|
snail family zinc finger 2 |
| chr1_+_144989309 | 0.17 |
ENST00000596396.1
|
AL590452.1
|
Uncharacterized protein |
| chr7_+_99699179 | 0.17 |
ENST00000438383.1
ENST00000429084.1 ENST00000359593.4 ENST00000439416.1 |
AP4M1
|
adaptor-related protein complex 4, mu 1 subunit |
| chr16_+_88704978 | 0.17 |
ENST00000244241.4
|
IL17C
|
interleukin 17C |
| chr1_-_31712401 | 0.16 |
ENST00000373736.2
|
NKAIN1
|
Na+/K+ transporting ATPase interacting 1 |
| chr6_+_31795506 | 0.16 |
ENST00000375650.3
|
HSPA1B
|
heat shock 70kDa protein 1B |
| chr17_+_4981535 | 0.16 |
ENST00000318833.3
|
ZFP3
|
ZFP3 zinc finger protein |
| chr12_-_53228079 | 0.16 |
ENST00000330553.5
|
KRT79
|
keratin 79 |
| chr7_+_99699280 | 0.16 |
ENST00000421755.1
|
AP4M1
|
adaptor-related protein complex 4, mu 1 subunit |
| chr7_-_8301768 | 0.15 |
ENST00000265577.7
|
ICA1
|
islet cell autoantigen 1, 69kDa |
| chr1_-_156675535 | 0.15 |
ENST00000368221.1
|
CRABP2
|
cellular retinoic acid binding protein 2 |
| chr12_-_62586543 | 0.15 |
ENST00000416284.3
|
FAM19A2
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A2 |
| chr2_+_46524537 | 0.15 |
ENST00000263734.3
|
EPAS1
|
endothelial PAS domain protein 1 |
| chr22_-_31063782 | 0.15 |
ENST00000404885.1
ENST00000403268.1 ENST00000407308.1 ENST00000342474.4 ENST00000334679.3 |
DUSP18
|
dual specificity phosphatase 18 |
| chr1_+_168250194 | 0.15 |
ENST00000367821.3
|
TBX19
|
T-box 19 |
| chr8_-_144512576 | 0.15 |
ENST00000333480.2
|
MAFA
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog A |
| chr8_+_22435762 | 0.15 |
ENST00000456545.1
|
PDLIM2
|
PDZ and LIM domain 2 (mystique) |
| chr19_-_9968816 | 0.15 |
ENST00000590841.1
|
OLFM2
|
olfactomedin 2 |
| chrX_-_11445856 | 0.15 |
ENST00000380736.1
|
ARHGAP6
|
Rho GTPase activating protein 6 |
| chr1_+_155051305 | 0.14 |
ENST00000368408.3
|
EFNA3
|
ephrin-A3 |
| chr17_+_12693001 | 0.14 |
ENST00000262444.9
|
ARHGAP44
|
Rho GTPase activating protein 44 |
| chr19_-_18391708 | 0.14 |
ENST00000600972.1
|
JUND
|
jun D proto-oncogene |
| chr2_-_128051670 | 0.14 |
ENST00000493187.2
|
ERCC3
|
excision repair cross-complementing rodent repair deficiency, complementation group 3 |
| chrX_+_151806637 | 0.14 |
ENST00000370306.2
|
GABRQ
|
gamma-aminobutyric acid (GABA) A receptor, theta |
| chr21_+_44313375 | 0.14 |
ENST00000354250.2
ENST00000340344.4 |
NDUFV3
|
NADH dehydrogenase (ubiquinone) flavoprotein 3, 10kDa |
| chr17_+_39382900 | 0.14 |
ENST00000377721.3
ENST00000455970.2 |
KRTAP9-2
|
keratin associated protein 9-2 |
| chr1_-_160990886 | 0.14 |
ENST00000537746.1
|
F11R
|
F11 receptor |
| chr6_+_96463840 | 0.14 |
ENST00000302103.5
|
FUT9
|
fucosyltransferase 9 (alpha (1,3) fucosyltransferase) |
| chr7_+_130794846 | 0.13 |
ENST00000421797.2
|
MKLN1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr2_-_225811747 | 0.13 |
ENST00000409592.3
|
DOCK10
|
dedicator of cytokinesis 10 |
| chr2_+_30670127 | 0.13 |
ENST00000540623.1
ENST00000476038.1 |
LCLAT1
|
lysocardiolipin acyltransferase 1 |
| chrX_-_118827333 | 0.13 |
ENST00000360156.7
ENST00000354228.4 ENST00000489216.1 ENST00000354416.3 ENST00000394610.1 ENST00000343984.5 |
SEPT6
|
septin 6 |
| chr16_-_4465886 | 0.13 |
ENST00000539968.1
|
CORO7
|
coronin 7 |
| chr7_-_99698338 | 0.13 |
ENST00000354230.3
ENST00000425308.1 |
MCM7
|
minichromosome maintenance complex component 7 |
| chr7_+_128470431 | 0.13 |
ENST00000325888.8
ENST00000346177.6 |
FLNC
|
filamin C, gamma |
| chrX_-_33146477 | 0.13 |
ENST00000378677.2
|
DMD
|
dystrophin |
| chr4_+_6271558 | 0.13 |
ENST00000503569.1
ENST00000226760.1 |
WFS1
|
Wolfram syndrome 1 (wolframin) |
| chr7_+_35756092 | 0.13 |
ENST00000458087.3
|
AC018647.3
|
AC018647.3 |
| chr6_+_34204642 | 0.13 |
ENST00000347617.6
ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1
|
high mobility group AT-hook 1 |
| chr2_-_179672142 | 0.13 |
ENST00000342992.6
ENST00000360870.5 ENST00000460472.2 ENST00000589042.1 ENST00000591111.1 ENST00000342175.6 ENST00000359218.5 |
TTN
|
titin |
| chr16_-_69419473 | 0.13 |
ENST00000566750.1
|
TERF2
|
telomeric repeat binding factor 2 |
| chr17_+_7462103 | 0.13 |
ENST00000396545.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr1_+_230203010 | 0.13 |
ENST00000541865.1
|
GALNT2
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-T2) |
| chr1_+_26737253 | 0.12 |
ENST00000326279.6
|
LIN28A
|
lin-28 homolog A (C. elegans) |
| chr5_+_150827143 | 0.12 |
ENST00000243389.3
ENST00000517945.1 ENST00000521925.1 |
SLC36A1
|
solute carrier family 36 (proton/amino acid symporter), member 1 |
| chr3_-_128207349 | 0.12 |
ENST00000487848.1
|
GATA2
|
GATA binding protein 2 |
| chr4_+_81951957 | 0.12 |
ENST00000282701.2
|
BMP3
|
bone morphogenetic protein 3 |
| chr6_+_41888926 | 0.12 |
ENST00000230340.4
|
BYSL
|
bystin-like |
| chr1_-_11120057 | 0.12 |
ENST00000376957.2
|
SRM
|
spermidine synthase |
| chr8_-_49834299 | 0.12 |
ENST00000396822.1
|
SNAI2
|
snail family zinc finger 2 |
| chr7_-_25702669 | 0.12 |
ENST00000446840.1
|
AC003090.1
|
AC003090.1 |
| chr16_-_69418649 | 0.12 |
ENST00000566257.1
|
TERF2
|
telomeric repeat binding factor 2 |
| chr16_+_4606347 | 0.12 |
ENST00000444310.4
|
C16orf96
|
chromosome 16 open reading frame 96 |
| chr7_-_8302164 | 0.12 |
ENST00000447326.1
ENST00000406470.2 |
ICA1
|
islet cell autoantigen 1, 69kDa |
| chr19_+_18794470 | 0.12 |
ENST00000321949.8
ENST00000338797.6 |
CRTC1
|
CREB regulated transcription coactivator 1 |
| chr1_-_201391149 | 0.12 |
ENST00000555948.1
ENST00000556362.1 |
TNNI1
|
troponin I type 1 (skeletal, slow) |
| chr4_-_186456766 | 0.12 |
ENST00000284771.6
|
PDLIM3
|
PDZ and LIM domain 3 |
| chr7_-_2883928 | 0.12 |
ENST00000275364.3
|
GNA12
|
guanine nucleotide binding protein (G protein) alpha 12 |
| chr6_+_12717892 | 0.11 |
ENST00000379350.1
|
PHACTR1
|
phosphatase and actin regulator 1 |
| chr2_+_28113583 | 0.11 |
ENST00000344773.2
ENST00000379624.1 ENST00000342045.2 ENST00000379632.2 ENST00000361704.2 |
BRE
|
brain and reproductive organ-expressed (TNFRSF1A modulator) |
| chr19_-_39881777 | 0.11 |
ENST00000595564.1
ENST00000221265.3 |
PAF1
|
Paf1, RNA polymerase II associated factor, homolog (S. cerevisiae) |
| chr12_-_56326402 | 0.11 |
ENST00000547925.1
|
WIBG
|
within bgcn homolog (Drosophila) |
| chr19_-_42916499 | 0.11 |
ENST00000601189.1
ENST00000599211.1 |
LIPE
|
lipase, hormone-sensitive |
| chr7_-_8301682 | 0.11 |
ENST00000396675.3
ENST00000430867.1 |
ICA1
|
islet cell autoantigen 1, 69kDa |
| chr9_+_133710453 | 0.10 |
ENST00000318560.5
|
ABL1
|
c-abl oncogene 1, non-receptor tyrosine kinase |
| chr11_+_118958689 | 0.10 |
ENST00000535253.1
ENST00000392841.1 |
HMBS
|
hydroxymethylbilane synthase |
| chr17_+_7461849 | 0.10 |
ENST00000338784.4
|
TNFSF13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr1_-_24741525 | 0.10 |
ENST00000374409.1
|
STPG1
|
sperm-tail PG-rich repeat containing 1 |
| chr11_-_65626797 | 0.10 |
ENST00000525451.2
|
CFL1
|
cofilin 1 (non-muscle) |
| chr19_+_50936142 | 0.10 |
ENST00000357701.5
|
MYBPC2
|
myosin binding protein C, fast type |
| chr12_+_50017327 | 0.10 |
ENST00000261897.1
|
PRPF40B
|
PRP40 pre-mRNA processing factor 40 homolog B (S. cerevisiae) |
| chr17_+_74732889 | 0.10 |
ENST00000591864.1
|
MFSD11
|
major facilitator superfamily domain containing 11 |
| chr17_-_39222131 | 0.10 |
ENST00000394015.2
|
KRTAP2-4
|
keratin associated protein 2-4 |
| chr5_+_140868717 | 0.10 |
ENST00000252087.1
|
PCDHGC5
|
protocadherin gamma subfamily C, 5 |
| chr2_-_176033066 | 0.10 |
ENST00000437522.1
|
ATF2
|
activating transcription factor 2 |
| chr4_-_177116772 | 0.09 |
ENST00000280191.2
|
SPATA4
|
spermatogenesis associated 4 |
| chr17_+_39405939 | 0.09 |
ENST00000334109.2
|
KRTAP9-4
|
keratin associated protein 9-4 |
| chr12_-_57006882 | 0.09 |
ENST00000551996.1
|
BAZ2A
|
bromodomain adjacent to zinc finger domain, 2A |
| chr6_+_46761118 | 0.09 |
ENST00000230588.4
|
MEP1A
|
meprin A, alpha (PABA peptide hydrolase) |
| chr9_-_139258159 | 0.09 |
ENST00000371739.3
|
DNLZ
|
DNL-type zinc finger |
| chr11_+_93754513 | 0.09 |
ENST00000315765.9
|
HEPHL1
|
hephaestin-like 1 |
| chr16_+_2286726 | 0.09 |
ENST00000382437.4
ENST00000569184.1 |
DNASE1L2
|
deoxyribonuclease I-like 2 |
| chrX_-_153775426 | 0.09 |
ENST00000393562.2
|
G6PD
|
glucose-6-phosphate dehydrogenase |
| chr7_-_82792215 | 0.08 |
ENST00000333891.9
ENST00000423517.2 |
PCLO
|
piccolo presynaptic cytomatrix protein |
| chr7_-_8302207 | 0.08 |
ENST00000407906.1
|
ICA1
|
islet cell autoantigen 1, 69kDa |
| chr1_-_154842741 | 0.08 |
ENST00000271915.4
|
KCNN3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3 |
| chr21_-_36260980 | 0.08 |
ENST00000344691.4
ENST00000358356.5 |
RUNX1
|
runt-related transcription factor 1 |
| chr19_-_3786253 | 0.08 |
ENST00000585778.1
|
MATK
|
megakaryocyte-associated tyrosine kinase |
| chr7_-_78400598 | 0.08 |
ENST00000536571.1
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr12_+_56325812 | 0.08 |
ENST00000394147.1
ENST00000551156.1 ENST00000553783.1 ENST00000557080.1 ENST00000432422.3 ENST00000556001.1 |
DGKA
|
diacylglycerol kinase, alpha 80kDa |
| chr19_-_10230540 | 0.08 |
ENST00000589454.1
|
EIF3G
|
eukaryotic translation initiation factor 3, subunit G |
| chr11_-_65626753 | 0.08 |
ENST00000526975.1
ENST00000531413.1 |
CFL1
|
cofilin 1 (non-muscle) |
| chr1_-_111991850 | 0.08 |
ENST00000411751.2
|
WDR77
|
WD repeat domain 77 |
| chr17_-_7145106 | 0.08 |
ENST00000577035.1
|
GABARAP
|
GABA(A) receptor-associated protein |
| chr1_+_66999268 | 0.08 |
ENST00000371039.1
ENST00000424320.1 |
SGIP1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr2_-_50574856 | 0.08 |
ENST00000342183.5
|
NRXN1
|
neurexin 1 |
| chr19_-_18632861 | 0.08 |
ENST00000262809.4
|
ELL
|
elongation factor RNA polymerase II |
| chr1_-_42384343 | 0.07 |
ENST00000372584.1
|
HIVEP3
|
human immunodeficiency virus type I enhancer binding protein 3 |
| chr7_+_93535817 | 0.07 |
ENST00000248572.5
|
GNGT1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr4_-_90757364 | 0.07 |
ENST00000508895.1
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr2_-_220436248 | 0.07 |
ENST00000265318.4
|
OBSL1
|
obscurin-like 1 |
| chr19_-_40324255 | 0.07 |
ENST00000593685.1
ENST00000600611.1 |
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr19_-_40791211 | 0.07 |
ENST00000579047.1
|
AKT2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr17_-_34890617 | 0.07 |
ENST00000586886.1
ENST00000585719.1 ENST00000585818.1 |
MYO19
|
myosin XIX |
| chr20_-_9819479 | 0.07 |
ENST00000378423.1
ENST00000353224.5 |
PAK7
|
p21 protein (Cdc42/Rac)-activated kinase 7 |
| chr14_-_71276211 | 0.07 |
ENST00000381250.4
ENST00000555993.2 |
MAP3K9
|
mitogen-activated protein kinase kinase kinase 9 |
| chr6_+_36238237 | 0.07 |
ENST00000457797.1
ENST00000394571.2 |
PNPLA1
|
patatin-like phospholipase domain containing 1 |
| chr19_+_39687596 | 0.07 |
ENST00000339852.4
|
NCCRP1
|
non-specific cytotoxic cell receptor protein 1 homolog (zebrafish) |
| chr3_-_50374869 | 0.07 |
ENST00000327761.3
|
RASSF1
|
Ras association (RalGDS/AF-6) domain family member 1 |
| chr11_-_30038490 | 0.07 |
ENST00000328224.6
|
KCNA4
|
potassium voltage-gated channel, shaker-related subfamily, member 4 |
| chr13_-_41635512 | 0.07 |
ENST00000405737.2
|
ELF1
|
E74-like factor 1 (ets domain transcription factor) |
| chr12_+_32654965 | 0.07 |
ENST00000472289.1
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr18_-_45663666 | 0.07 |
ENST00000535628.2
|
ZBTB7C
|
zinc finger and BTB domain containing 7C |
| chr19_-_10230562 | 0.07 |
ENST00000587146.1
ENST00000588709.1 ENST00000253108.4 |
EIF3G
|
eukaryotic translation initiation factor 3, subunit G |
| chr17_-_31204124 | 0.07 |
ENST00000579584.1
ENST00000318217.5 ENST00000583621.1 |
MYO1D
|
myosin ID |
| chr16_-_74808710 | 0.07 |
ENST00000219368.3
ENST00000544337.1 |
FA2H
|
fatty acid 2-hydroxylase |
Network of associatons between targets according to the STRING database.
First level regulatory network of FIGLA
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.5 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.3 | 0.8 | GO:0021776 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.1 | 0.5 | GO:1903517 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) negative regulation of single strand break repair(GO:1903517) negative regulation of beta-galactosidase activity(GO:1903770) telomere single strand break repair(GO:1903823) negative regulation of telomere single strand break repair(GO:1903824) |
| 0.1 | 0.3 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.1 | 0.5 | GO:0085032 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.1 | 0.4 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.1 | 0.3 | GO:0070426 | positive regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070426) positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070434) |
| 0.1 | 0.2 | GO:0002232 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) |
| 0.1 | 0.8 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 2.2 | GO:0097062 | dendritic spine maintenance(GO:0097062) |
| 0.1 | 0.2 | GO:0032903 | viral protein processing(GO:0019082) regulation of nerve growth factor production(GO:0032903) negative regulation of nerve growth factor production(GO:0032904) dibasic protein processing(GO:0090472) |
| 0.1 | 0.3 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.2 | GO:1902723 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.0 | 0.4 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.1 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.0 | 0.1 | GO:0035854 | regulation of primitive erythrocyte differentiation(GO:0010725) eosinophil fate commitment(GO:0035854) |
| 0.0 | 0.2 | GO:0032425 | positive regulation of mismatch repair(GO:0032425) |
| 0.0 | 0.2 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 0.6 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.1 | GO:1990051 | activation of protein kinase C activity(GO:1990051) |
| 0.0 | 0.3 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:2000314 | regulation of neural crest formation(GO:0090299) negative regulation of neural crest formation(GO:0090301) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.0 | 0.5 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.1 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.3 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.1 | GO:0090402 | senescence-associated heterochromatin focus assembly(GO:0035986) oncogene-induced cell senescence(GO:0090402) |
| 0.0 | 0.1 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.0 | 0.6 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.1 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.1 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.0 | 0.1 | GO:0003335 | corneocyte development(GO:0003335) |
| 0.0 | 0.3 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 | 0.1 | GO:1901895 | negative regulation of calcium-transporting ATPase activity(GO:1901895) |
| 0.0 | 0.1 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) |
| 0.0 | 0.2 | GO:0097396 | response to interleukin-17(GO:0097396) cellular response to interleukin-17(GO:0097398) |
| 0.0 | 0.1 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.1 | GO:1902303 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.0 | 0.2 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.2 | GO:0038129 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 0.2 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.5 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.3 | GO:0021554 | optic nerve development(GO:0021554) |
| 0.0 | 0.2 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.4 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.1 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.2 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 0.1 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.1 | GO:0031630 | regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031630) |
| 0.0 | 0.5 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.1 | GO:0051622 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.0 | 0.1 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.2 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.1 | GO:0035524 | L-alanine transport(GO:0015808) proline transmembrane transport(GO:0035524) |
| 0.0 | 0.1 | GO:0048747 | muscle fiber development(GO:0048747) |
| 0.0 | 0.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.1 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.1 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.0 | 0.1 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.1 | GO:1905247 | positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.0 | 0.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:0071486 | cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.0 | 0.1 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.9 | GO:0034667 | integrin alpha3-beta1 complex(GO:0034667) |
| 0.2 | 0.6 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.0 | 0.1 | GO:0005940 | septin ring(GO:0005940) septin collar(GO:0032173) |
| 0.0 | 0.2 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.4 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.3 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.5 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.1 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 1.6 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.6 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.1 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.0 | GO:0016935 | glycine-gated chloride channel complex(GO:0016935) |
| 0.0 | 0.8 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.1 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.1 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.1 | GO:0042025 | host cell nucleus(GO:0042025) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.1 | 0.5 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.1 | 0.8 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 0.2 | GO:0004968 | gonadotropin-releasing hormone receptor activity(GO:0004968) |
| 0.0 | 1.9 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.1 | GO:0004766 | spermidine synthase activity(GO:0004766) |
| 0.0 | 0.2 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.0 | 0.1 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.3 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.5 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.3 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.6 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.2 | GO:0043813 | phosphatidylinositol-3,5-bisphosphate 5-phosphatase activity(GO:0043813) |
| 0.0 | 0.4 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.0 | 0.1 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.3 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0032558 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.0 | 0.1 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 0.2 | GO:0035252 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.2 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.1 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.0 | 0.8 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.1 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0004515 | nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.1 | GO:0015180 | L-alanine transmembrane transporter activity(GO:0015180) L-proline transmembrane transporter activity(GO:0015193) alanine transmembrane transporter activity(GO:0022858) |
| 0.0 | 0.2 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.0 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.9 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.6 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.4 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.7 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.6 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.2 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 1.5 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.8 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.4 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.4 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |