Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
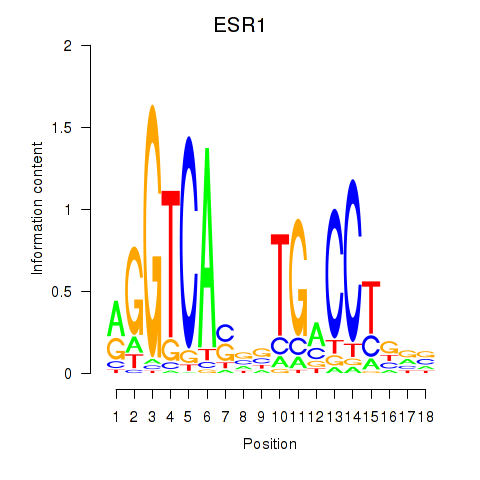
Results for ESR1
Z-value: 0.52
Transcription factors associated with ESR1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ESR1
|
ENSG00000091831.17 | estrogen receptor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ESR1 | hg19_v2_chr6_+_152130240_152130274 | -0.36 | 6.4e-01 | Click! |
Activity profile of ESR1 motif
Sorted Z-values of ESR1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_-_47352477 | 0.24 |
ENST00000593412.1
|
PRED62
|
Uncharacterized protein |
| chr8_-_144674284 | 0.20 |
ENST00000528519.1
|
EEF1D
|
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chr16_+_28857916 | 0.19 |
ENST00000563591.1
|
SH2B1
|
SH2B adaptor protein 1 |
| chrX_-_21776281 | 0.17 |
ENST00000379494.3
|
SMPX
|
small muscle protein, X-linked |
| chr19_+_19516561 | 0.17 |
ENST00000457895.2
|
GATAD2A
|
GATA zinc finger domain containing 2A |
| chr1_-_38157877 | 0.17 |
ENST00000477060.1
ENST00000491981.1 ENST00000488137.1 |
C1orf109
|
chromosome 1 open reading frame 109 |
| chr2_-_74779744 | 0.17 |
ENST00000409249.1
|
LOXL3
|
lysyl oxidase-like 3 |
| chr11_-_9482010 | 0.17 |
ENST00000596206.1
|
AC132192.1
|
LOC644656 protein; Uncharacterized protein |
| chr22_+_44427230 | 0.16 |
ENST00000444029.1
|
PARVB
|
parvin, beta |
| chr11_+_62439126 | 0.15 |
ENST00000377953.3
|
C11orf83
|
chromosome 11 open reading frame 83 |
| chr6_+_35265586 | 0.14 |
ENST00000542066.1
ENST00000316637.5 |
DEF6
|
differentially expressed in FDCP 6 homolog (mouse) |
| chr8_-_103136481 | 0.14 |
ENST00000524209.1
ENST00000517822.1 ENST00000523923.1 ENST00000521599.1 ENST00000521964.1 ENST00000311028.3 ENST00000518166.1 |
NCALD
|
neurocalcin delta |
| chr22_+_22723969 | 0.14 |
ENST00000390295.2
|
IGLV7-46
|
immunoglobulin lambda variable 7-46 (gene/pseudogene) |
| chr2_+_242750274 | 0.13 |
ENST00000405370.1
|
NEU4
|
sialidase 4 |
| chr2_+_131328402 | 0.13 |
ENST00000409793.1
ENST00000409982.1 |
AC140481.2
|
Uncharacterized protein |
| chr16_-_51185149 | 0.13 |
ENST00000566102.1
ENST00000541611.1 |
SALL1
|
spalt-like transcription factor 1 |
| chr12_-_124457163 | 0.13 |
ENST00000535556.1
|
CCDC92
|
coiled-coil domain containing 92 |
| chr9_-_21368075 | 0.12 |
ENST00000449498.1
|
IFNA13
|
interferon, alpha 13 |
| chr19_-_10613421 | 0.12 |
ENST00000393623.2
|
KEAP1
|
kelch-like ECH-associated protein 1 |
| chr16_-_88772670 | 0.12 |
ENST00000562544.1
|
RNF166
|
ring finger protein 166 |
| chr1_-_111970353 | 0.12 |
ENST00000369732.3
|
OVGP1
|
oviductal glycoprotein 1, 120kDa |
| chr7_+_140396946 | 0.11 |
ENST00000476470.1
ENST00000471136.1 |
NDUFB2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2, 8kDa |
| chr22_+_22676808 | 0.11 |
ENST00000390290.2
|
IGLV1-51
|
immunoglobulin lambda variable 1-51 |
| chr22_-_32034376 | 0.11 |
ENST00000431201.1
|
PISD
|
phosphatidylserine decarboxylase |
| chr17_+_65027509 | 0.11 |
ENST00000375684.1
|
AC005544.1
|
Uncharacterized protein |
| chr4_+_89300158 | 0.10 |
ENST00000502870.1
|
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr20_+_48429233 | 0.10 |
ENST00000417961.1
|
SLC9A8
|
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chr14_+_103995546 | 0.10 |
ENST00000299202.4
|
TRMT61A
|
tRNA methyltransferase 61 homolog A (S. cerevisiae) |
| chr22_+_24199113 | 0.10 |
ENST00000405847.1
|
SLC2A11
|
solute carrier family 2 (facilitated glucose transporter), member 11 |
| chr15_+_75335604 | 0.10 |
ENST00000563393.1
|
PPCDC
|
phosphopantothenoylcysteine decarboxylase |
| chr8_+_144640499 | 0.10 |
ENST00000525721.1
ENST00000534018.1 |
GSDMD
|
gasdermin D |
| chr16_-_19725899 | 0.10 |
ENST00000567367.1
|
KNOP1
|
lysine-rich nucleolar protein 1 |
| chr22_-_32034409 | 0.10 |
ENST00000429683.1
|
PISD
|
phosphatidylserine decarboxylase |
| chr5_+_32710736 | 0.10 |
ENST00000415685.2
|
NPR3
|
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr11_-_86383650 | 0.10 |
ENST00000526944.1
ENST00000530335.1 ENST00000543262.1 ENST00000524826.1 |
ME3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial |
| chr2_+_27719697 | 0.10 |
ENST00000264717.2
ENST00000424318.2 |
GCKR
|
glucokinase (hexokinase 4) regulator |
| chr19_-_11639910 | 0.10 |
ENST00000588998.1
ENST00000586149.1 |
ECSIT
|
ECSIT signalling integrator |
| chr4_-_89079817 | 0.09 |
ENST00000505480.1
|
ABCG2
|
ATP-binding cassette, sub-family G (WHITE), member 2 |
| chr12_+_123874589 | 0.09 |
ENST00000437502.1
|
SETD8
|
SET domain containing (lysine methyltransferase) 8 |
| chr7_+_140396756 | 0.09 |
ENST00000460088.1
ENST00000472695.1 |
NDUFB2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2, 8kDa |
| chr12_+_100967420 | 0.09 |
ENST00000266754.5
ENST00000547754.1 |
GAS2L3
|
growth arrest-specific 2 like 3 |
| chr5_+_154135453 | 0.09 |
ENST00000517616.1
ENST00000518892.1 |
LARP1
|
La ribonucleoprotein domain family, member 1 |
| chr7_-_99569468 | 0.09 |
ENST00000419575.1
|
AZGP1
|
alpha-2-glycoprotein 1, zinc-binding |
| chr14_+_52780998 | 0.09 |
ENST00000557436.1
|
PTGER2
|
prostaglandin E receptor 2 (subtype EP2), 53kDa |
| chr1_+_55446465 | 0.09 |
ENST00000371268.3
|
TMEM61
|
transmembrane protein 61 |
| chr14_-_23791484 | 0.09 |
ENST00000594872.1
|
AL049829.1
|
Uncharacterized protein |
| chr20_+_42295745 | 0.09 |
ENST00000396863.4
ENST00000217026.4 |
MYBL2
|
v-myb avian myeloblastosis viral oncogene homolog-like 2 |
| chr20_+_49411523 | 0.09 |
ENST00000371608.2
|
BCAS4
|
breast carcinoma amplified sequence 4 |
| chr8_-_123793048 | 0.09 |
ENST00000607710.1
|
RP11-44N11.2
|
RP11-44N11.2 |
| chr19_-_10445399 | 0.09 |
ENST00000592945.1
|
ICAM3
|
intercellular adhesion molecule 3 |
| chr16_+_28857957 | 0.09 |
ENST00000567536.1
|
SH2B1
|
SH2B adaptor protein 1 |
| chr7_+_79763271 | 0.09 |
ENST00000442586.1
|
GNAI1
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
| chr11_+_134146635 | 0.09 |
ENST00000431683.2
|
GLB1L3
|
galactosidase, beta 1-like 3 |
| chr19_+_1261106 | 0.09 |
ENST00000588411.1
|
CIRBP
|
cold inducible RNA binding protein |
| chr19_-_13068012 | 0.08 |
ENST00000316939.1
|
GADD45GIP1
|
growth arrest and DNA-damage-inducible, gamma interacting protein 1 |
| chr1_-_16763685 | 0.08 |
ENST00000540400.1
|
SPATA21
|
spermatogenesis associated 21 |
| chr17_-_39677971 | 0.08 |
ENST00000393976.2
|
KRT15
|
keratin 15 |
| chr19_-_11639931 | 0.08 |
ENST00000592312.1
ENST00000590480.1 ENST00000585318.1 ENST00000252440.7 ENST00000417981.2 ENST00000270517.7 |
ECSIT
|
ECSIT signalling integrator |
| chr6_+_41196052 | 0.08 |
ENST00000341495.2
|
TREML4
|
triggering receptor expressed on myeloid cells-like 4 |
| chr3_-_52488048 | 0.08 |
ENST00000232975.3
|
TNNC1
|
troponin C type 1 (slow) |
| chr19_-_59010565 | 0.08 |
ENST00000594786.1
|
SLC27A5
|
solute carrier family 27 (fatty acid transporter), member 5 |
| chr11_+_8704748 | 0.08 |
ENST00000526562.1
ENST00000525981.1 |
RPL27A
|
ribosomal protein L27a |
| chr8_+_144640477 | 0.08 |
ENST00000262580.4
|
GSDMD
|
gasdermin D |
| chr22_+_30821732 | 0.08 |
ENST00000355143.4
|
MTFP1
|
mitochondrial fission process 1 |
| chr2_+_233243233 | 0.08 |
ENST00000392027.2
|
ALPP
|
alkaline phosphatase, placental |
| chr11_+_47270475 | 0.08 |
ENST00000481889.2
ENST00000436778.1 ENST00000531660.1 ENST00000407404.1 |
NR1H3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr14_+_103995503 | 0.08 |
ENST00000389749.4
|
TRMT61A
|
tRNA methyltransferase 61 homolog A (S. cerevisiae) |
| chr11_-_62439012 | 0.08 |
ENST00000532208.1
ENST00000377954.2 ENST00000415855.2 ENST00000431002.2 ENST00000354588.3 |
C11orf48
|
chromosome 11 open reading frame 48 |
| chr8_-_100905363 | 0.08 |
ENST00000524245.1
|
COX6C
|
cytochrome c oxidase subunit VIc |
| chrX_-_106959631 | 0.08 |
ENST00000486554.1
ENST00000372390.4 |
TSC22D3
|
TSC22 domain family, member 3 |
| chr19_-_51869592 | 0.08 |
ENST00000596253.1
ENST00000309244.4 |
ETFB
|
electron-transfer-flavoprotein, beta polypeptide |
| chr19_-_36001386 | 0.08 |
ENST00000461300.1
|
DMKN
|
dermokine |
| chr7_+_149535455 | 0.08 |
ENST00000223210.4
ENST00000460379.1 |
ZNF862
|
zinc finger protein 862 |
| chr19_+_58790314 | 0.08 |
ENST00000196548.5
ENST00000608843.1 |
ZNF8
ZNF8
|
Zinc finger protein 8 zinc finger protein 8 |
| chr11_-_86383370 | 0.08 |
ENST00000526834.1
ENST00000359636.2 |
ME3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial |
| chr19_-_49567124 | 0.07 |
ENST00000301411.3
|
NTF4
|
neurotrophin 4 |
| chr4_+_128982490 | 0.07 |
ENST00000394288.3
ENST00000432347.2 ENST00000264584.5 ENST00000441387.1 ENST00000427266.1 ENST00000354456.3 |
LARP1B
|
La ribonucleoprotein domain family, member 1B |
| chr3_-_47554791 | 0.07 |
ENST00000449409.1
ENST00000414236.1 ENST00000444760.1 ENST00000439305.1 |
ELP6
|
elongator acetyltransferase complex subunit 6 |
| chrX_+_153770421 | 0.07 |
ENST00000369609.5
ENST00000369607.1 |
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr17_+_73629500 | 0.07 |
ENST00000375215.3
|
SMIM5
|
small integral membrane protein 5 |
| chr19_+_8740061 | 0.07 |
ENST00000593792.1
|
CTD-2586B10.1
|
CTD-2586B10.1 |
| chr1_-_20306909 | 0.07 |
ENST00000375111.3
ENST00000400520.3 |
PLA2G2A
|
phospholipase A2, group IIA (platelets, synovial fluid) |
| chr10_+_134210672 | 0.07 |
ENST00000305233.5
ENST00000368609.4 |
PWWP2B
|
PWWP domain containing 2B |
| chr17_-_73285293 | 0.07 |
ENST00000582778.1
ENST00000581988.1 ENST00000579207.1 ENST00000583332.1 ENST00000416858.2 ENST00000442286.2 ENST00000580151.1 ENST00000580994.1 ENST00000584438.1 ENST00000320362.3 ENST00000580273.1 |
SLC25A19
|
solute carrier family 25 (mitochondrial thiamine pyrophosphate carrier), member 19 |
| chr19_-_2090131 | 0.07 |
ENST00000591326.1
|
MOB3A
|
MOB kinase activator 3A |
| chr8_-_144691718 | 0.07 |
ENST00000377579.3
ENST00000433751.1 ENST00000220966.6 |
PYCRL
|
pyrroline-5-carboxylate reductase-like |
| chr15_+_101389945 | 0.07 |
ENST00000561231.1
ENST00000559331.1 ENST00000558254.1 |
RP11-66B24.2
|
RP11-66B24.2 |
| chr20_+_3801162 | 0.07 |
ENST00000379573.2
ENST00000379567.2 ENST00000455742.1 ENST00000246041.2 |
AP5S1
|
adaptor-related protein complex 5, sigma 1 subunit |
| chr9_-_127177703 | 0.07 |
ENST00000259457.3
ENST00000536392.1 ENST00000441097.1 |
PSMB7
|
proteasome (prosome, macropain) subunit, beta type, 7 |
| chr3_+_38017264 | 0.07 |
ENST00000436654.1
|
CTDSPL
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
| chr16_+_28858004 | 0.07 |
ENST00000322610.8
|
SH2B1
|
SH2B adaptor protein 1 |
| chr1_+_145726886 | 0.07 |
ENST00000443667.1
|
PDZK1
|
PDZ domain containing 1 |
| chr11_+_47270436 | 0.07 |
ENST00000395397.3
ENST00000405576.1 |
NR1H3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr19_+_47840346 | 0.07 |
ENST00000600626.1
|
C5AR2
|
complement component 5a receptor 2 |
| chr1_+_245133656 | 0.07 |
ENST00000366521.3
|
EFCAB2
|
EF-hand calcium binding domain 2 |
| chr20_+_48429356 | 0.07 |
ENST00000361573.2
ENST00000541138.1 ENST00000539601.1 |
SLC9A8
|
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chr16_+_50730910 | 0.07 |
ENST00000300589.2
|
NOD2
|
nucleotide-binding oligomerization domain containing 2 |
| chr4_-_89080362 | 0.07 |
ENST00000503830.1
|
ABCG2
|
ATP-binding cassette, sub-family G (WHITE), member 2 |
| chr19_-_29218538 | 0.07 |
ENST00000592347.1
ENST00000586528.1 |
AC005307.3
|
AC005307.3 |
| chr19_+_7562431 | 0.07 |
ENST00000361664.2
|
C19orf45
|
chromosome 19 open reading frame 45 |
| chr7_-_6066183 | 0.07 |
ENST00000422786.1
|
EIF2AK1
|
eukaryotic translation initiation factor 2-alpha kinase 1 |
| chr12_+_16064258 | 0.07 |
ENST00000524480.1
ENST00000531803.1 ENST00000532964.1 |
DERA
|
deoxyribose-phosphate aldolase (putative) |
| chr16_-_67427389 | 0.06 |
ENST00000562206.1
ENST00000290942.5 ENST00000393957.2 |
TPPP3
|
tubulin polymerization-promoting protein family member 3 |
| chr9_+_129987488 | 0.06 |
ENST00000446764.1
|
GARNL3
|
GTPase activating Rap/RanGAP domain-like 3 |
| chr2_+_219264466 | 0.06 |
ENST00000273062.2
|
CTDSP1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1 |
| chr12_-_56123444 | 0.06 |
ENST00000546457.1
ENST00000549117.1 |
CD63
|
CD63 molecule |
| chrX_-_1571759 | 0.06 |
ENST00000381317.3
ENST00000416733.2 |
ASMTL
|
acetylserotonin O-methyltransferase-like |
| chr1_+_6684918 | 0.06 |
ENST00000054650.4
|
THAP3
|
THAP domain containing, apoptosis associated protein 3 |
| chr1_-_21620877 | 0.06 |
ENST00000527991.1
|
ECE1
|
endothelin converting enzyme 1 |
| chr3_+_15469058 | 0.06 |
ENST00000432764.2
|
EAF1
|
ELL associated factor 1 |
| chr1_+_52521928 | 0.06 |
ENST00000489308.2
|
BTF3L4
|
basic transcription factor 3-like 4 |
| chr1_+_44444865 | 0.06 |
ENST00000372324.1
|
B4GALT2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr1_+_1217305 | 0.06 |
ENST00000470022.1
|
SCNN1D
|
sodium channel, non-voltage-gated 1, delta subunit |
| chr9_-_130637244 | 0.06 |
ENST00000373156.1
|
AK1
|
adenylate kinase 1 |
| chr1_+_245133606 | 0.06 |
ENST00000447569.2
|
EFCAB2
|
EF-hand calcium binding domain 2 |
| chr8_-_100905850 | 0.06 |
ENST00000520271.1
ENST00000522940.1 ENST00000523016.1 ENST00000517682.2 ENST00000297564.2 |
COX6C
|
cytochrome c oxidase subunit VIc |
| chr16_+_50308028 | 0.06 |
ENST00000566761.2
|
ADCY7
|
adenylate cyclase 7 |
| chr4_-_155511887 | 0.06 |
ENST00000302053.3
ENST00000403106.3 |
FGA
|
fibrinogen alpha chain |
| chr5_+_149109825 | 0.06 |
ENST00000360453.4
ENST00000394320.3 ENST00000309241.5 |
PPARGC1B
|
peroxisome proliferator-activated receptor gamma, coactivator 1 beta |
| chr19_-_14586125 | 0.06 |
ENST00000292513.3
|
PTGER1
|
prostaglandin E receptor 1 (subtype EP1), 42kDa |
| chr1_-_167906020 | 0.06 |
ENST00000458574.1
|
MPC2
|
mitochondrial pyruvate carrier 2 |
| chr15_+_91418918 | 0.06 |
ENST00000560824.1
|
FURIN
|
furin (paired basic amino acid cleaving enzyme) |
| chr11_+_844406 | 0.06 |
ENST00000397404.1
|
TSPAN4
|
tetraspanin 4 |
| chr19_+_7985198 | 0.06 |
ENST00000221573.6
ENST00000595637.1 |
SNAPC2
|
small nuclear RNA activating complex, polypeptide 2, 45kDa |
| chr1_+_19923454 | 0.06 |
ENST00000602662.1
ENST00000602293.1 ENST00000322753.6 |
MINOS1-NBL1
MINOS1
|
MINOS1-NBL1 readthrough mitochondrial inner membrane organizing system 1 |
| chr5_-_139726181 | 0.06 |
ENST00000507104.1
ENST00000230990.6 |
HBEGF
|
heparin-binding EGF-like growth factor |
| chr14_-_100841794 | 0.06 |
ENST00000556295.1
ENST00000554820.1 |
WARS
|
tryptophanyl-tRNA synthetase |
| chr17_-_26989136 | 0.06 |
ENST00000247020.4
|
SDF2
|
stromal cell-derived factor 2 |
| chr3_-_15106747 | 0.06 |
ENST00000449354.2
ENST00000444840.2 ENST00000253686.2 |
MRPS25
|
mitochondrial ribosomal protein S25 |
| chr19_-_13213954 | 0.06 |
ENST00000590974.1
|
LYL1
|
lymphoblastic leukemia derived sequence 1 |
| chr19_+_1908013 | 0.06 |
ENST00000454697.1
|
ADAT3
|
adenosine deaminase, tRNA-specific 3 |
| chr8_-_41522779 | 0.06 |
ENST00000522231.1
ENST00000314214.8 ENST00000348036.4 ENST00000457297.1 ENST00000522543.1 |
ANK1
|
ankyrin 1, erythrocytic |
| chr22_+_41258250 | 0.06 |
ENST00000544094.1
|
XPNPEP3
|
X-prolyl aminopeptidase (aminopeptidase P) 3, putative |
| chr10_+_112257596 | 0.06 |
ENST00000369583.3
|
DUSP5
|
dual specificity phosphatase 5 |
| chr3_-_176914963 | 0.06 |
ENST00000450267.1
ENST00000431674.1 |
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1 |
| chr19_+_56111680 | 0.06 |
ENST00000301073.3
|
ZNF524
|
zinc finger protein 524 |
| chr12_-_31158902 | 0.06 |
ENST00000544329.1
ENST00000418254.2 ENST00000222396.5 |
RP11-551L14.4
|
RP11-551L14.4 |
| chr19_-_10614386 | 0.06 |
ENST00000171111.5
|
KEAP1
|
kelch-like ECH-associated protein 1 |
| chr1_+_245133278 | 0.06 |
ENST00000366522.2
|
EFCAB2
|
EF-hand calcium binding domain 2 |
| chr12_-_120806960 | 0.05 |
ENST00000257552.2
|
MSI1
|
musashi RNA-binding protein 1 |
| chr1_+_203097407 | 0.05 |
ENST00000367235.1
|
ADORA1
|
adenosine A1 receptor |
| chr9_-_120177342 | 0.05 |
ENST00000361209.2
|
ASTN2
|
astrotactin 2 |
| chr2_-_74776586 | 0.05 |
ENST00000420535.1
|
LOXL3
|
lysyl oxidase-like 3 |
| chr11_-_1780261 | 0.05 |
ENST00000427721.1
|
RP11-295K3.1
|
RP11-295K3.1 |
| chr4_+_25915896 | 0.05 |
ENST00000514384.1
|
SMIM20
|
small integral membrane protein 20 |
| chr11_+_1295809 | 0.05 |
ENST00000598274.1
|
AC136297.1
|
Uncharacterized protein |
| chr9_+_101984577 | 0.05 |
ENST00000223641.4
|
SEC61B
|
Sec61 beta subunit |
| chr3_+_37903432 | 0.05 |
ENST00000443503.2
|
CTDSPL
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
| chr16_+_75182376 | 0.05 |
ENST00000570010.1
ENST00000568079.1 ENST00000464850.1 ENST00000332307.4 ENST00000393430.2 |
ZFP1
|
ZFP1 zinc finger protein |
| chr8_+_40018977 | 0.05 |
ENST00000520487.1
|
RP11-470M17.2
|
RP11-470M17.2 |
| chr17_-_40346477 | 0.05 |
ENST00000593209.1
ENST00000587427.1 ENST00000588352.1 ENST00000414034.3 ENST00000590249.1 |
GHDC
|
GH3 domain containing |
| chr17_-_27507377 | 0.05 |
ENST00000531253.1
|
MYO18A
|
myosin XVIIIA |
| chr16_+_89686991 | 0.05 |
ENST00000393092.3
|
DPEP1
|
dipeptidase 1 (renal) |
| chr9_-_95896550 | 0.05 |
ENST00000375446.4
|
NINJ1
|
ninjurin 1 |
| chr15_-_76603727 | 0.05 |
ENST00000560595.1
ENST00000433983.2 ENST00000559386.1 ENST00000559602.1 ENST00000560726.1 ENST00000557943.1 |
ETFA
|
electron-transfer-flavoprotein, alpha polypeptide |
| chr10_+_131265443 | 0.05 |
ENST00000306010.7
|
MGMT
|
O-6-methylguanine-DNA methyltransferase |
| chr16_+_29690358 | 0.05 |
ENST00000395384.4
ENST00000562473.1 |
QPRT
|
quinolinate phosphoribosyltransferase |
| chr1_+_38259459 | 0.05 |
ENST00000373045.6
|
MANEAL
|
mannosidase, endo-alpha-like |
| chr17_-_27507395 | 0.05 |
ENST00000354329.4
ENST00000527372.1 |
MYO18A
|
myosin XVIIIA |
| chr11_+_2397418 | 0.05 |
ENST00000530648.1
|
CD81
|
CD81 molecule |
| chr16_-_2581409 | 0.05 |
ENST00000567119.1
ENST00000565480.1 ENST00000382350.1 |
CEMP1
|
cementum protein 1 |
| chr16_+_2079637 | 0.05 |
ENST00000561844.1
|
SLC9A3R2
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 2 |
| chr5_-_175395283 | 0.05 |
ENST00000513482.1
ENST00000265097.4 |
THOC3
|
THO complex 3 |
| chr17_-_73840774 | 0.05 |
ENST00000207549.4
|
UNC13D
|
unc-13 homolog D (C. elegans) |
| chr4_-_89080003 | 0.05 |
ENST00000237612.3
|
ABCG2
|
ATP-binding cassette, sub-family G (WHITE), member 2 |
| chr22_+_39853258 | 0.05 |
ENST00000341184.6
|
MGAT3
|
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase |
| chr2_+_183982255 | 0.05 |
ENST00000455063.1
|
NUP35
|
nucleoporin 35kDa |
| chr17_+_55173933 | 0.05 |
ENST00000539273.1
|
AKAP1
|
A kinase (PRKA) anchor protein 1 |
| chr7_-_1609591 | 0.05 |
ENST00000288607.2
ENST00000404674.3 |
PSMG3
|
proteasome (prosome, macropain) assembly chaperone 3 |
| chr8_-_101571964 | 0.05 |
ENST00000520552.1
ENST00000521345.1 ENST00000523000.1 ENST00000335659.3 ENST00000358990.3 ENST00000519597.1 |
ANKRD46
|
ankyrin repeat domain 46 |
| chr17_-_79869340 | 0.05 |
ENST00000538936.2
|
PCYT2
|
phosphate cytidylyltransferase 2, ethanolamine |
| chr22_-_21579843 | 0.05 |
ENST00000405188.4
|
GGT2
|
gamma-glutamyltransferase 2 |
| chr11_-_503521 | 0.05 |
ENST00000534797.1
|
RNH1
|
ribonuclease/angiogenin inhibitor 1 |
| chr16_+_88772866 | 0.05 |
ENST00000453996.2
ENST00000312060.5 ENST00000378384.3 ENST00000567949.1 ENST00000564921.1 |
CTU2
|
cytosolic thiouridylase subunit 2 homolog (S. pombe) |
| chr6_-_44225231 | 0.05 |
ENST00000538577.1
ENST00000537814.1 ENST00000393810.1 ENST00000393812.3 |
SLC35B2
|
solute carrier family 35 (adenosine 3'-phospho 5'-phosphosulfate transporter), member B2 |
| chr1_+_179851999 | 0.05 |
ENST00000527391.1
|
TOR1AIP1
|
torsin A interacting protein 1 |
| chr20_+_56964169 | 0.05 |
ENST00000475243.1
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chrX_+_55101495 | 0.05 |
ENST00000374974.3
ENST00000374971.1 |
PAGE2B
|
P antigen family, member 2B |
| chr7_+_6071007 | 0.05 |
ENST00000409061.1
|
ANKRD61
|
ankyrin repeat domain 61 |
| chr7_+_140396465 | 0.05 |
ENST00000476279.1
ENST00000247866.4 ENST00000461457.1 ENST00000465506.1 ENST00000204307.5 ENST00000464566.1 |
NDUFB2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2, 8kDa |
| chr18_+_33552667 | 0.05 |
ENST00000333234.5
|
C18orf21
|
chromosome 18 open reading frame 21 |
| chr1_+_246729724 | 0.05 |
ENST00000366513.4
ENST00000366512.3 |
CNST
|
consortin, connexin sorting protein |
| chr20_+_44441626 | 0.05 |
ENST00000372568.4
|
UBE2C
|
ubiquitin-conjugating enzyme E2C |
| chr19_-_18995029 | 0.05 |
ENST00000596048.1
|
CERS1
|
ceramide synthase 1 |
| chr19_+_39936267 | 0.05 |
ENST00000359191.6
|
SUPT5H
|
suppressor of Ty 5 homolog (S. cerevisiae) |
| chr17_+_6347729 | 0.05 |
ENST00000572447.1
|
FAM64A
|
family with sequence similarity 64, member A |
| chr17_+_27052892 | 0.05 |
ENST00000579671.1
ENST00000579060.1 |
NEK8
|
NIMA-related kinase 8 |
| chr20_+_32581738 | 0.05 |
ENST00000333552.5
|
RALY
|
RALY heterogeneous nuclear ribonucleoprotein |
| chr19_+_39574945 | 0.05 |
ENST00000331256.5
|
PAPL
|
Iron/zinc purple acid phosphatase-like protein |
| chr11_+_111750206 | 0.05 |
ENST00000530214.1
ENST00000530799.1 |
C11orf1
|
chromosome 11 open reading frame 1 |
| chr19_+_39936186 | 0.05 |
ENST00000432763.2
ENST00000402194.2 ENST00000601515.1 |
SUPT5H
|
suppressor of Ty 5 homolog (S. cerevisiae) |
| chr11_-_75062829 | 0.05 |
ENST00000393505.4
|
ARRB1
|
arrestin, beta 1 |
| chr6_+_33168637 | 0.05 |
ENST00000374677.3
|
SLC39A7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr11_+_844067 | 0.05 |
ENST00000397406.1
ENST00000409543.2 ENST00000525201.1 |
TSPAN4
|
tetraspanin 4 |
| chr1_-_36916011 | 0.05 |
ENST00000356637.5
ENST00000354267.3 ENST00000235532.5 |
OSCP1
|
organic solute carrier partner 1 |
| chr16_-_28621298 | 0.05 |
ENST00000566189.1
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr14_+_77790901 | 0.05 |
ENST00000553586.1
ENST00000555583.1 |
GSTZ1
|
glutathione S-transferase zeta 1 |
| chr16_+_718147 | 0.05 |
ENST00000561929.1
|
RHOT2
|
ras homolog family member T2 |
| chr9_-_35619539 | 0.05 |
ENST00000396757.1
|
CD72
|
CD72 molecule |
| chr11_+_64008525 | 0.05 |
ENST00000449942.2
|
FKBP2
|
FK506 binding protein 2, 13kDa |
Network of associatons between targets according to the STRING database.
First level regulatory network of ESR1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:2000329 | peptidyl-lysine oxidation(GO:0018057) negative regulation of T-helper 17 cell lineage commitment(GO:2000329) |
| 0.0 | 0.1 | GO:2000360 | negative regulation of fertilization(GO:0060467) negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.0 | 0.1 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.0 | 0.1 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 0.0 | 0.1 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.0 | 0.1 | GO:0010160 | sensory organ boundary specification(GO:0008052) formation of organ boundary(GO:0010160) taste bud development(GO:0061193) |
| 0.0 | 0.2 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.1 | GO:0032900 | negative regulation of neurotrophin production(GO:0032900) |
| 0.0 | 0.1 | GO:0060584 | activation of MAPK activity involved in innate immune response(GO:0035419) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.0 | 0.1 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.0 | 0.1 | GO:0038178 | complement component C5a signaling pathway(GO:0038178) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.0 | 0.2 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.0 | 0.1 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.0 | 0.1 | GO:0016999 | antibiotic metabolic process(GO:0016999) |
| 0.0 | 0.0 | GO:1902559 | 3'-phosphoadenosine 5'-phosphosulfate transport(GO:0046963) 3'-phospho-5'-adenylyl sulfate transmembrane transport(GO:1902559) |
| 0.0 | 0.2 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 0.0 | 0.1 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.0 | GO:0072720 | cellular response to mycotoxin(GO:0036146) response to dithiothreitol(GO:0072720) |
| 0.0 | 0.1 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.2 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.1 | GO:0072752 | cellular response to rapamycin(GO:0072752) |
| 0.0 | 0.0 | GO:1902824 | cleavage furrow ingression(GO:0036090) lysosomal membrane organization(GO:0097212) positive regulation of late endosome to lysosome transport(GO:1902824) |
| 0.0 | 0.0 | GO:1903452 | regulation of G1 to G0 transition(GO:1903450) positive regulation of G1 to G0 transition(GO:1903452) |
| 0.0 | 0.1 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.1 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.0 | 0.0 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.1 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) |
| 0.0 | 0.1 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.0 | GO:0070894 | transposon integration(GO:0070893) regulation of transposon integration(GO:0070894) negative regulation of transposon integration(GO:0070895) |
| 0.0 | 0.1 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.0 | 0.0 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.0 | 0.0 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.0 | 0.0 | GO:0098968 | neurotransmitter receptor transport postsynaptic membrane to endosome(GO:0098968) |
| 0.0 | 0.1 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.0 | 0.1 | GO:0070235 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.1 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.0 | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.0 | 0.1 | GO:1901091 | regulation of protein tetramerization(GO:1901090) negative regulation of protein tetramerization(GO:1901091) regulation of protein homotetramerization(GO:1901093) negative regulation of protein homotetramerization(GO:1901094) |
| 0.0 | 0.0 | GO:0070845 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.0 | 0.1 | GO:0010816 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.0 | 0.2 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.1 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 0.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.1 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.0 | 0.0 | GO:0043227 | membrane-bounded organelle(GO:0043227) |
| 0.0 | 0.0 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.0 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.2 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.0 | 0.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.1 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.0 | 0.0 | GO:0035579 | specific granule membrane(GO:0035579) |
| 0.0 | 0.0 | GO:0001534 | radial spoke(GO:0001534) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0016426 | tRNA (adenine) methyltransferase activity(GO:0016426) tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.0 | 0.2 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.0 | 0.1 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 0.0 | 0.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0004878 | complement component C5a receptor activity(GO:0004878) |
| 0.0 | 0.1 | GO:0052595 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.1 | GO:0052794 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.0 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 0.1 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.0 | 0.2 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.0 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.0 | 0.2 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.0 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.0 | 0.1 | GO:1990450 | linear polyubiquitin binding(GO:1990450) |
| 0.0 | 0.1 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) |
| 0.0 | 0.1 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.0 | 0.1 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.1 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 0.1 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.1 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.0 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.0 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.0 | GO:0050571 | 1,5-anhydro-D-fructose reductase activity(GO:0050571) |
| 0.0 | 0.0 | GO:0047291 | neolactotetraosylceramide alpha-2,3-sialyltransferase activity(GO:0004513) lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |