Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
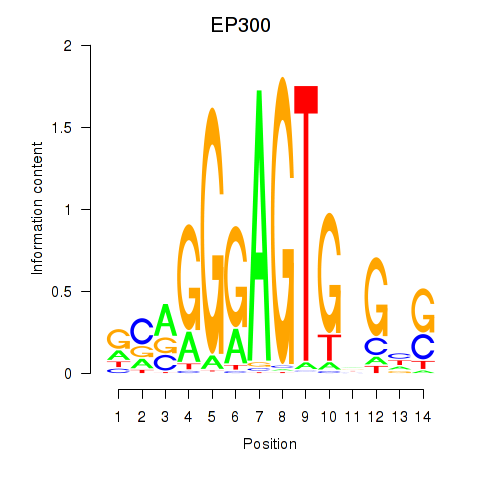
Results for EP300
Z-value: 0.28
Transcription factors associated with EP300
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EP300
|
ENSG00000100393.9 | E1A binding protein p300 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EP300 | hg19_v2_chr22_+_41487711_41487798 | -0.68 | 3.2e-01 | Click! |
Activity profile of EP300 motif
Sorted Z-values of EP300 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_39832563 | 0.22 |
ENST00000599274.1
|
CTC-246B18.10
|
CTC-246B18.10 |
| chr6_-_35888858 | 0.12 |
ENST00000507909.1
|
SRPK1
|
SRSF protein kinase 1 |
| chr2_+_87808725 | 0.10 |
ENST00000413202.1
|
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr2_-_32390801 | 0.10 |
ENST00000608489.1
|
RP11-563N4.1
|
RP11-563N4.1 |
| chr2_+_25016282 | 0.09 |
ENST00000260662.1
|
CENPO
|
centromere protein O |
| chr1_+_211432700 | 0.09 |
ENST00000452621.2
|
RCOR3
|
REST corepressor 3 |
| chr12_-_124118151 | 0.08 |
ENST00000534960.1
|
EIF2B1
|
eukaryotic translation initiation factor 2B, subunit 1 alpha, 26kDa |
| chr3_+_238969 | 0.08 |
ENST00000421198.1
|
CHL1
|
cell adhesion molecule L1-like |
| chr16_-_30204987 | 0.08 |
ENST00000569282.1
ENST00000567436.1 |
BOLA2B
|
bolA family member 2B |
| chr11_+_46722368 | 0.07 |
ENST00000311764.2
|
ZNF408
|
zinc finger protein 408 |
| chr4_-_110736505 | 0.07 |
ENST00000609440.1
|
RP11-602N24.3
|
RP11-602N24.3 |
| chr14_-_23284703 | 0.07 |
ENST00000555911.1
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr11_-_62389621 | 0.07 |
ENST00000531383.1
ENST00000265471.5 |
B3GAT3
|
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) |
| chr2_+_20866424 | 0.07 |
ENST00000272224.3
|
GDF7
|
growth differentiation factor 7 |
| chr11_+_63870660 | 0.07 |
ENST00000246841.3
|
FLRT1
|
fibronectin leucine rich transmembrane protein 1 |
| chr7_-_944631 | 0.06 |
ENST00000453175.2
|
ADAP1
|
ArfGAP with dual PH domains 1 |
| chr12_+_52463751 | 0.06 |
ENST00000336854.4
ENST00000550604.1 ENST00000553049.1 ENST00000548915.1 |
C12orf44
|
chromosome 12 open reading frame 44 |
| chr17_+_72427477 | 0.06 |
ENST00000342648.5
ENST00000481232.1 |
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr22_-_31688381 | 0.06 |
ENST00000487265.2
|
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr22_+_37257015 | 0.06 |
ENST00000447071.1
ENST00000248899.6 ENST00000397147.4 |
NCF4
|
neutrophil cytosolic factor 4, 40kDa |
| chr11_+_2421718 | 0.06 |
ENST00000380996.5
ENST00000333256.6 ENST00000380992.1 ENST00000437110.1 ENST00000435795.1 |
TSSC4
|
tumor suppressing subtransferable candidate 4 |
| chr5_-_138725594 | 0.06 |
ENST00000302125.8
|
MZB1
|
marginal zone B and B1 cell-specific protein |
| chr22_+_23229960 | 0.06 |
ENST00000526893.1
ENST00000532223.2 ENST00000531372.1 |
IGLL5
|
immunoglobulin lambda-like polypeptide 5 |
| chr17_-_7080227 | 0.05 |
ENST00000574330.1
|
ASGR1
|
asialoglycoprotein receptor 1 |
| chr11_-_62389449 | 0.05 |
ENST00000534026.1
|
B3GAT3
|
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) |
| chr2_+_220379052 | 0.05 |
ENST00000347842.3
ENST00000358078.4 |
ASIC4
|
acid-sensing (proton-gated) ion channel family member 4 |
| chr16_+_4784458 | 0.05 |
ENST00000590191.1
|
C16orf71
|
chromosome 16 open reading frame 71 |
| chr6_+_3000218 | 0.05 |
ENST00000380441.1
ENST00000380455.4 ENST00000380454.4 |
NQO2
|
NAD(P)H dehydrogenase, quinone 2 |
| chr11_+_62623544 | 0.05 |
ENST00000377890.2
ENST00000377891.2 ENST00000377889.2 |
SLC3A2
|
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr5_-_138725560 | 0.05 |
ENST00000412103.2
ENST00000457570.2 |
MZB1
|
marginal zone B and B1 cell-specific protein |
| chr6_+_3000195 | 0.05 |
ENST00000338130.2
|
NQO2
|
NAD(P)H dehydrogenase, quinone 2 |
| chrX_+_23685653 | 0.05 |
ENST00000379331.3
|
PRDX4
|
peroxiredoxin 4 |
| chr9_+_127631399 | 0.05 |
ENST00000259477.6
|
ARPC5L
|
actin related protein 2/3 complex, subunit 5-like |
| chr19_+_1261106 | 0.05 |
ENST00000588411.1
|
CIRBP
|
cold inducible RNA binding protein |
| chr9_-_127703333 | 0.05 |
ENST00000373555.4
|
GOLGA1
|
golgin A1 |
| chr2_+_110371905 | 0.05 |
ENST00000356454.3
|
SOWAHC
|
sosondowah ankyrin repeat domain family member C |
| chr19_+_16296191 | 0.05 |
ENST00000589852.1
ENST00000263384.7 ENST00000588367.1 ENST00000587351.1 |
FAM32A
|
family with sequence similarity 32, member A |
| chr1_-_47697387 | 0.05 |
ENST00000371884.2
|
TAL1
|
T-cell acute lymphocytic leukemia 1 |
| chr4_+_56814968 | 0.05 |
ENST00000422247.2
|
CEP135
|
centrosomal protein 135kDa |
| chr2_+_217277137 | 0.05 |
ENST00000430374.1
ENST00000357276.4 ENST00000444508.1 |
SMARCAL1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a-like 1 |
| chr22_-_37640456 | 0.05 |
ENST00000405484.1
ENST00000441619.1 ENST00000406508.1 |
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr11_+_62623621 | 0.04 |
ENST00000535296.1
|
SLC3A2
|
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr1_+_109656579 | 0.04 |
ENST00000526264.1
ENST00000369939.3 |
KIAA1324
|
KIAA1324 |
| chr17_-_19265982 | 0.04 |
ENST00000268841.6
ENST00000261499.4 ENST00000575478.1 |
B9D1
|
B9 protein domain 1 |
| chr11_+_62623512 | 0.04 |
ENST00000377892.1
|
SLC3A2
|
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr12_+_53693812 | 0.04 |
ENST00000549488.1
|
C12orf10
|
chromosome 12 open reading frame 10 |
| chr19_+_19627026 | 0.04 |
ENST00000608404.1
ENST00000555938.1 ENST00000503283.1 ENST00000512771.3 ENST00000428459.2 |
YJEFN3
CTC-260F20.3
NDUFA13
|
YjeF N-terminal domain containing 3 Uncharacterized protein NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 13 |
| chr6_-_30684898 | 0.04 |
ENST00000422266.1
ENST00000416571.1 |
MDC1
|
mediator of DNA-damage checkpoint 1 |
| chr16_-_87799505 | 0.04 |
ENST00000353170.5
ENST00000561825.1 ENST00000270583.5 ENST00000562261.1 ENST00000347925.5 |
KLHDC4
|
kelch domain containing 4 |
| chr11_+_64879317 | 0.04 |
ENST00000526809.1
ENST00000279263.7 ENST00000524986.1 ENST00000534371.1 ENST00000540748.1 ENST00000525385.1 ENST00000345348.5 ENST00000531321.1 ENST00000529414.1 ENST00000526085.1 ENST00000530750.1 |
TM7SF2
|
transmembrane 7 superfamily member 2 |
| chr9_-_72287191 | 0.04 |
ENST00000265381.4
|
APBA1
|
amyloid beta (A4) precursor protein-binding, family A, member 1 |
| chr17_-_76837499 | 0.04 |
ENST00000592275.1
|
USP36
|
ubiquitin specific peptidase 36 |
| chr17_-_79995553 | 0.04 |
ENST00000581584.1
ENST00000577712.1 ENST00000582900.1 ENST00000579155.1 ENST00000306869.2 |
DCXR
|
dicarbonyl/L-xylulose reductase |
| chr10_-_99161033 | 0.04 |
ENST00000315563.6
ENST00000370992.4 ENST00000414986.1 |
RRP12
|
ribosomal RNA processing 12 homolog (S. cerevisiae) |
| chr19_-_49955050 | 0.04 |
ENST00000262265.5
|
PIH1D1
|
PIH1 domain containing 1 |
| chr16_-_66968055 | 0.04 |
ENST00000568572.1
|
FAM96B
|
family with sequence similarity 96, member B |
| chr18_+_9334786 | 0.04 |
ENST00000581641.1
|
TWSG1
|
twisted gastrulation BMP signaling modulator 1 |
| chr1_-_33366931 | 0.04 |
ENST00000373463.3
ENST00000329151.5 |
TMEM54
|
transmembrane protein 54 |
| chr17_-_56065540 | 0.04 |
ENST00000583932.1
|
VEZF1
|
vascular endothelial zinc finger 1 |
| chr2_+_74154032 | 0.04 |
ENST00000356837.6
|
DGUOK
|
deoxyguanosine kinase |
| chr19_+_49617609 | 0.04 |
ENST00000221459.2
ENST00000486217.2 |
LIN7B
|
lin-7 homolog B (C. elegans) |
| chr19_-_44809121 | 0.04 |
ENST00000591609.1
ENST00000589799.1 ENST00000291182.4 ENST00000589248.1 |
ZNF235
|
zinc finger protein 235 |
| chr12_+_7072354 | 0.04 |
ENST00000537269.1
|
U47924.27
|
U47924.27 |
| chr6_-_150039170 | 0.04 |
ENST00000458696.2
ENST00000392273.3 |
LATS1
|
large tumor suppressor kinase 1 |
| chr15_-_82338460 | 0.04 |
ENST00000558133.1
ENST00000329713.4 |
MEX3B
|
mex-3 RNA binding family member B |
| chr2_-_233877912 | 0.04 |
ENST00000264051.3
|
NGEF
|
neuronal guanine nucleotide exchange factor |
| chr18_+_77439775 | 0.04 |
ENST00000299543.7
ENST00000075430.7 |
CTDP1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1 |
| chr15_+_74908228 | 0.04 |
ENST00000566126.1
|
CLK3
|
CDC-like kinase 3 |
| chr19_-_17186229 | 0.04 |
ENST00000253669.5
ENST00000448593.2 |
HAUS8
|
HAUS augmin-like complex, subunit 8 |
| chr17_-_19266045 | 0.04 |
ENST00000395616.3
|
B9D1
|
B9 protein domain 1 |
| chr22_-_20104700 | 0.04 |
ENST00000439169.2
ENST00000445045.1 ENST00000404751.3 ENST00000252136.7 ENST00000403707.3 |
TRMT2A
|
tRNA methyltransferase 2 homolog A (S. cerevisiae) |
| chr16_-_66968265 | 0.04 |
ENST00000567511.1
ENST00000422424.2 |
FAM96B
|
family with sequence similarity 96, member B |
| chr11_-_62389577 | 0.04 |
ENST00000534715.1
|
B3GAT3
|
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) |
| chr11_+_236540 | 0.04 |
ENST00000532097.1
|
PSMD13
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 13 |
| chr19_-_10613421 | 0.04 |
ENST00000393623.2
|
KEAP1
|
kelch-like ECH-associated protein 1 |
| chr6_-_30685214 | 0.04 |
ENST00000425072.1
|
MDC1
|
mediator of DNA-damage checkpoint 1 |
| chr11_+_237016 | 0.04 |
ENST00000352303.5
|
PSMD13
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 13 |
| chr17_-_74099772 | 0.04 |
ENST00000411744.2
ENST00000332065.5 |
EXOC7
|
exocyst complex component 7 |
| chr6_-_33290580 | 0.04 |
ENST00000446511.1
ENST00000446403.1 ENST00000414083.2 ENST00000266000.6 ENST00000374542.5 |
DAXX
|
death-domain associated protein |
| chr16_+_68279207 | 0.04 |
ENST00000413021.2
ENST00000565744.1 ENST00000219345.5 |
PLA2G15
|
phospholipase A2, group XV |
| chr10_-_104192405 | 0.04 |
ENST00000369937.4
|
CUEDC2
|
CUE domain containing 2 |
| chr10_-_64028466 | 0.04 |
ENST00000395265.1
ENST00000373789.3 ENST00000395260.3 |
RTKN2
|
rhotekin 2 |
| chr15_+_76196234 | 0.04 |
ENST00000540507.1
ENST00000565036.1 ENST00000569054.1 |
FBXO22
|
F-box protein 22 |
| chr1_+_153651078 | 0.04 |
ENST00000368680.3
|
NPR1
|
natriuretic peptide receptor A/guanylate cyclase A (atrionatriuretic peptide receptor A) |
| chr17_-_37844267 | 0.04 |
ENST00000579146.1
ENST00000378011.4 ENST00000429199.2 ENST00000300658.4 |
PGAP3
|
post-GPI attachment to proteins 3 |
| chr17_+_72426891 | 0.04 |
ENST00000392627.1
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr1_-_247495045 | 0.04 |
ENST00000294753.4
ENST00000366498.2 |
ZNF496
|
zinc finger protein 496 |
| chr17_-_74099795 | 0.04 |
ENST00000406660.3
ENST00000335146.7 ENST00000405575.4 ENST00000589210.1 ENST00000607838.1 |
EXOC7
|
exocyst complex component 7 |
| chr20_+_61340179 | 0.04 |
ENST00000370501.3
|
NTSR1
|
neurotensin receptor 1 (high affinity) |
| chr3_-_135915146 | 0.03 |
ENST00000473093.1
|
MSL2
|
male-specific lethal 2 homolog (Drosophila) |
| chr17_+_49230897 | 0.03 |
ENST00000393196.3
ENST00000336097.3 ENST00000480143.1 ENST00000511355.1 ENST00000013034.3 ENST00000393198.3 ENST00000608447.1 ENST00000393193.2 ENST00000376392.6 ENST00000555572.1 |
NME1
NME1-NME2
NME2
|
NME/NM23 nucleoside diphosphate kinase 1 NME1-NME2 readthrough NME/NM23 nucleoside diphosphate kinase 2 |
| chrX_-_54522558 | 0.03 |
ENST00000375135.3
|
FGD1
|
FYVE, RhoGEF and PH domain containing 1 |
| chr5_+_443268 | 0.03 |
ENST00000512944.1
|
EXOC3
|
exocyst complex component 3 |
| chr17_+_43238438 | 0.03 |
ENST00000593138.1
ENST00000586681.1 |
HEXIM2
|
hexamethylene bis-acetamide inducible 2 |
| chr12_+_121416489 | 0.03 |
ENST00000541395.1
ENST00000544413.1 |
HNF1A
|
HNF1 homeobox A |
| chr5_-_1345199 | 0.03 |
ENST00000320895.5
|
CLPTM1L
|
CLPTM1-like |
| chr18_-_5238525 | 0.03 |
ENST00000581170.1
ENST00000579933.1 ENST00000581067.1 |
RP11-835E18.5
LINC00526
|
RP11-835E18.5 long intergenic non-protein coding RNA 526 |
| chr19_-_36705763 | 0.03 |
ENST00000591473.1
|
ZNF565
|
zinc finger protein 565 |
| chr19_-_49955096 | 0.03 |
ENST00000595550.1
|
PIH1D1
|
PIH1 domain containing 1 |
| chr20_-_30311703 | 0.03 |
ENST00000450273.1
ENST00000456404.1 ENST00000420488.1 ENST00000439267.1 |
BCL2L1
|
BCL2-like 1 |
| chr17_-_19265855 | 0.03 |
ENST00000440841.1
ENST00000395615.1 ENST00000461069.2 |
B9D1
|
B9 protein domain 1 |
| chr19_-_11039188 | 0.03 |
ENST00000588347.1
|
YIPF2
|
Yip1 domain family, member 2 |
| chr1_+_116519223 | 0.03 |
ENST00000369502.1
|
SLC22A15
|
solute carrier family 22, member 15 |
| chr12_-_49525175 | 0.03 |
ENST00000336023.5
ENST00000550367.1 ENST00000552984.1 ENST00000547476.1 |
TUBA1B
|
tubulin, alpha 1b |
| chr6_-_32083106 | 0.03 |
ENST00000442721.1
|
TNXB
|
tenascin XB |
| chr6_-_107436473 | 0.03 |
ENST00000369042.1
|
BEND3
|
BEN domain containing 3 |
| chr16_+_1832902 | 0.03 |
ENST00000262302.9
ENST00000563136.1 ENST00000565987.1 ENST00000543305.1 ENST00000568287.1 ENST00000565134.1 |
NUBP2
|
nucleotide binding protein 2 |
| chr1_+_155657737 | 0.03 |
ENST00000471642.2
ENST00000471214.1 |
DAP3
|
death associated protein 3 |
| chr8_+_106330920 | 0.03 |
ENST00000407775.2
|
ZFPM2
|
zinc finger protein, FOG family member 2 |
| chr6_+_20403997 | 0.03 |
ENST00000535432.1
|
E2F3
|
E2F transcription factor 3 |
| chr17_+_4843679 | 0.03 |
ENST00000576229.1
|
RNF167
|
ring finger protein 167 |
| chr11_+_2397402 | 0.03 |
ENST00000475945.2
|
CD81
|
CD81 molecule |
| chr20_+_55043647 | 0.03 |
ENST00000023939.4
ENST00000395881.3 ENST00000357348.5 ENST00000449062.1 ENST00000435342.2 |
RTFDC1
|
replication termination factor 2 domain containing 1 |
| chr11_-_62342375 | 0.03 |
ENST00000378019.3
|
EEF1G
|
eukaryotic translation elongation factor 1 gamma |
| chr17_-_74137374 | 0.03 |
ENST00000322957.6
|
FOXJ1
|
forkhead box J1 |
| chr16_+_2083265 | 0.03 |
ENST00000565855.1
ENST00000566198.1 |
SLC9A3R2
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 2 |
| chr14_-_61190754 | 0.03 |
ENST00000216513.4
|
SIX4
|
SIX homeobox 4 |
| chr17_-_7518145 | 0.03 |
ENST00000250113.7
ENST00000571597.1 |
FXR2
|
fragile X mental retardation, autosomal homolog 2 |
| chr19_+_19626531 | 0.03 |
ENST00000507754.4
|
NDUFA13
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 13 |
| chr1_-_43855444 | 0.03 |
ENST00000372455.4
|
MED8
|
mediator complex subunit 8 |
| chrX_-_153599578 | 0.03 |
ENST00000360319.4
ENST00000344736.4 |
FLNA
|
filamin A, alpha |
| chr10_+_104178946 | 0.03 |
ENST00000432590.1
|
FBXL15
|
F-box and leucine-rich repeat protein 15 |
| chr17_-_26989136 | 0.03 |
ENST00000247020.4
|
SDF2
|
stromal cell-derived factor 2 |
| chr5_-_443239 | 0.03 |
ENST00000408966.2
|
C5orf55
|
chromosome 5 open reading frame 55 |
| chr19_+_45844018 | 0.03 |
ENST00000585434.1
|
KLC3
|
kinesin light chain 3 |
| chr20_+_47835884 | 0.03 |
ENST00000371764.4
|
DDX27
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 27 |
| chr9_-_139760717 | 0.03 |
ENST00000371648.4
|
EDF1
|
endothelial differentiation-related factor 1 |
| chr4_+_110736659 | 0.03 |
ENST00000394631.3
ENST00000226796.6 |
GAR1
|
GAR1 ribonucleoprotein |
| chr19_-_10514184 | 0.03 |
ENST00000589629.1
ENST00000222005.2 |
CDC37
|
cell division cycle 37 |
| chr22_-_19109901 | 0.03 |
ENST00000545799.1
ENST00000537045.1 ENST00000263196.7 |
DGCR2
|
DiGeorge syndrome critical region gene 2 |
| chr12_-_105629852 | 0.03 |
ENST00000551662.1
ENST00000553097.1 |
APPL2
|
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
| chr6_+_26087646 | 0.03 |
ENST00000309234.6
|
HFE
|
hemochromatosis |
| chr12_+_121416340 | 0.03 |
ENST00000257555.6
ENST00000400024.2 |
HNF1A
|
HNF1 homeobox A |
| chr6_-_149969829 | 0.03 |
ENST00000367411.2
|
KATNA1
|
katanin p60 (ATPase containing) subunit A 1 |
| chr1_+_206643806 | 0.03 |
ENST00000537984.1
|
IKBKE
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon |
| chr15_+_42120283 | 0.03 |
ENST00000542534.2
ENST00000397299.4 ENST00000408047.1 ENST00000431823.1 ENST00000382448.4 ENST00000342159.4 |
PLA2G4B
JMJD7
JMJD7-PLA2G4B
|
phospholipase A2, group IVB (cytosolic) jumonji domain containing 7 JMJD7-PLA2G4B readthrough |
| chr17_+_37844331 | 0.03 |
ENST00000578199.1
ENST00000406381.2 |
ERBB2
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 |
| chr1_+_3541543 | 0.03 |
ENST00000378344.2
ENST00000344579.5 |
TPRG1L
|
tumor protein p63 regulated 1-like |
| chr13_-_111358504 | 0.03 |
ENST00000257347.4
|
CARS2
|
cysteinyl-tRNA synthetase 2, mitochondrial (putative) |
| chr9_+_131218698 | 0.03 |
ENST00000434106.3
ENST00000546203.1 ENST00000446274.1 ENST00000421776.2 ENST00000432065.2 |
ODF2
|
outer dense fiber of sperm tails 2 |
| chrX_+_134166333 | 0.03 |
ENST00000257013.7
|
FAM127A
|
family with sequence similarity 127, member A |
| chr6_-_41703296 | 0.03 |
ENST00000373033.1
|
TFEB
|
transcription factor EB |
| chr16_+_718086 | 0.03 |
ENST00000315082.4
ENST00000563134.1 |
RHOT2
|
ras homolog family member T2 |
| chr19_-_4670345 | 0.03 |
ENST00000599630.1
ENST00000262947.3 |
C19orf10
|
chromosome 19 open reading frame 10 |
| chr16_+_29984962 | 0.03 |
ENST00000308893.4
|
TAOK2
|
TAO kinase 2 |
| chr16_+_69373323 | 0.03 |
ENST00000254940.5
|
NIP7
|
NIP7, nucleolar pre-rRNA processing protein |
| chr12_-_49393092 | 0.03 |
ENST00000421952.2
|
DDN
|
dendrin |
| chr1_-_160549235 | 0.03 |
ENST00000368054.3
ENST00000368048.3 ENST00000311224.4 ENST00000368051.3 ENST00000534968.1 |
CD84
|
CD84 molecule |
| chr6_-_28226984 | 0.03 |
ENST00000423974.2
|
ZKSCAN4
|
zinc finger with KRAB and SCAN domains 4 |
| chr19_-_50083803 | 0.03 |
ENST00000391853.3
ENST00000339093.3 |
NOSIP
|
nitric oxide synthase interacting protein |
| chr10_+_99400443 | 0.03 |
ENST00000370631.3
|
PI4K2A
|
phosphatidylinositol 4-kinase type 2 alpha |
| chr3_-_49066811 | 0.03 |
ENST00000442157.1
ENST00000326739.4 |
IMPDH2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2 |
| chr12_+_53693466 | 0.03 |
ENST00000267103.5
ENST00000548632.1 |
C12orf10
|
chromosome 12 open reading frame 10 |
| chr20_+_54987305 | 0.03 |
ENST00000371336.3
ENST00000434344.1 |
CASS4
|
Cas scaffolding protein family member 4 |
| chr11_-_107582775 | 0.03 |
ENST00000305991.2
|
SLN
|
sarcolipin |
| chr5_-_1344999 | 0.03 |
ENST00000320927.6
|
CLPTM1L
|
CLPTM1-like |
| chr16_+_4475806 | 0.03 |
ENST00000262375.6
ENST00000355296.4 ENST00000431375.2 ENST00000574895.1 |
DNAJA3
|
DnaJ (Hsp40) homolog, subfamily A, member 3 |
| chr4_+_8201091 | 0.02 |
ENST00000382521.3
ENST00000245105.3 ENST00000457650.2 ENST00000539824.1 |
SH3TC1
|
SH3 domain and tetratricopeptide repeats 1 |
| chr6_+_31865552 | 0.02 |
ENST00000469372.1
ENST00000497706.1 |
C2
|
complement component 2 |
| chr1_+_11866270 | 0.02 |
ENST00000376497.3
ENST00000376487.3 ENST00000376496.3 |
CLCN6
|
chloride channel, voltage-sensitive 6 |
| chr1_-_19229014 | 0.02 |
ENST00000538839.1
ENST00000290597.5 |
ALDH4A1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr2_+_27193480 | 0.02 |
ENST00000233121.2
ENST00000405074.3 |
MAPRE3
|
microtubule-associated protein, RP/EB family, member 3 |
| chr15_+_90895471 | 0.02 |
ENST00000354377.3
ENST00000379090.5 |
ZNF774
|
zinc finger protein 774 |
| chr11_-_18610246 | 0.02 |
ENST00000379387.4
ENST00000541984.1 |
UEVLD
|
UEV and lactate/malate dehyrogenase domains |
| chr19_+_10362882 | 0.02 |
ENST00000393733.2
ENST00000588502.1 |
MRPL4
|
mitochondrial ribosomal protein L4 |
| chr5_+_119799927 | 0.02 |
ENST00000407149.2
ENST00000379551.2 |
PRR16
|
proline rich 16 |
| chr9_+_140149625 | 0.02 |
ENST00000343053.4
|
NELFB
|
negative elongation factor complex member B |
| chr8_-_33424636 | 0.02 |
ENST00000256257.1
|
RNF122
|
ring finger protein 122 |
| chrX_+_54947229 | 0.02 |
ENST00000442098.1
ENST00000430420.1 ENST00000453081.1 ENST00000173898.7 ENST00000319167.8 ENST00000375022.4 ENST00000399736.1 ENST00000440072.1 ENST00000420798.2 ENST00000431115.1 ENST00000440759.1 ENST00000375041.2 |
TRO
|
trophinin |
| chr19_-_49314269 | 0.02 |
ENST00000545387.2
ENST00000316273.6 ENST00000402551.1 ENST00000598162.1 ENST00000599246.1 |
BCAT2
|
branched chain amino-acid transaminase 2, mitochondrial |
| chr22_+_20105012 | 0.02 |
ENST00000331821.3
ENST00000411892.1 |
RANBP1
|
RAN binding protein 1 |
| chr17_-_7137582 | 0.02 |
ENST00000575756.1
ENST00000575458.1 |
DVL2
|
dishevelled segment polarity protein 2 |
| chr12_+_58148842 | 0.02 |
ENST00000266643.5
|
MARCH9
|
membrane-associated ring finger (C3HC4) 9 |
| chr8_-_33330595 | 0.02 |
ENST00000524021.1
ENST00000335589.3 |
FUT10
|
fucosyltransferase 10 (alpha (1,3) fucosyltransferase) |
| chr2_-_204400013 | 0.02 |
ENST00000374489.2
ENST00000374488.2 ENST00000308091.4 ENST00000453034.1 ENST00000420371.1 |
RAPH1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
| chr11_-_2924720 | 0.02 |
ENST00000455942.2
|
SLC22A18AS
|
solute carrier family 22 (organic cation transporter), member 18 antisense |
| chr14_-_23284675 | 0.02 |
ENST00000555959.1
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr11_+_64323098 | 0.02 |
ENST00000301891.4
|
SLC22A11
|
solute carrier family 22 (organic anion/urate transporter), member 11 |
| chr17_+_27895045 | 0.02 |
ENST00000580183.2
ENST00000578749.1 ENST00000582829.2 |
TP53I13
|
tumor protein p53 inducible protein 13 |
| chr10_-_12238071 | 0.02 |
ENST00000491614.1
ENST00000537776.1 |
NUDT5
|
nudix (nucleoside diphosphate linked moiety X)-type motif 5 |
| chr1_+_95582881 | 0.02 |
ENST00000370203.4
ENST00000456991.1 |
TMEM56
|
transmembrane protein 56 |
| chr6_+_26087509 | 0.02 |
ENST00000397022.3
ENST00000353147.5 ENST00000352392.4 ENST00000349999.4 ENST00000317896.7 ENST00000357618.5 ENST00000470149.1 ENST00000336625.8 ENST00000461397.1 ENST00000488199.1 |
HFE
|
hemochromatosis |
| chr9_-_139760737 | 0.02 |
ENST00000371649.1
ENST00000224073.1 |
EDF1
|
endothelial differentiation-related factor 1 |
| chr9_-_133814455 | 0.02 |
ENST00000448616.1
|
FIBCD1
|
fibrinogen C domain containing 1 |
| chr19_+_10362577 | 0.02 |
ENST00000592514.1
ENST00000307422.5 ENST00000253099.6 ENST00000590150.1 ENST00000590669.1 |
MRPL4
|
mitochondrial ribosomal protein L4 |
| chr17_-_79269067 | 0.02 |
ENST00000288439.5
ENST00000374759.3 |
SLC38A10
|
solute carrier family 38, member 10 |
| chrX_+_153770421 | 0.02 |
ENST00000369609.5
ENST00000369607.1 |
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr7_-_2354099 | 0.02 |
ENST00000222990.3
|
SNX8
|
sorting nexin 8 |
| chr2_+_233415363 | 0.02 |
ENST00000409514.1
ENST00000409098.1 ENST00000409495.1 ENST00000409167.3 ENST00000409322.1 ENST00000409394.1 |
EIF4E2
|
eukaryotic translation initiation factor 4E family member 2 |
| chr11_+_65819802 | 0.02 |
ENST00000528302.1
ENST00000322535.6 ENST00000524627.1 ENST00000533595.1 ENST00000530322.1 |
SF3B2
|
splicing factor 3b, subunit 2, 145kDa |
| chr20_+_43104508 | 0.02 |
ENST00000262605.4
ENST00000372904.3 |
TTPAL
|
tocopherol (alpha) transfer protein-like |
| chr16_-_69373396 | 0.02 |
ENST00000562595.1
ENST00000562081.1 ENST00000306875.4 |
COG8
|
component of oligomeric golgi complex 8 |
| chr14_+_52327350 | 0.02 |
ENST00000555472.1
ENST00000556766.1 |
GNG2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr5_+_42423872 | 0.02 |
ENST00000230882.4
ENST00000357703.3 |
GHR
|
growth hormone receptor |
| chr1_-_8938736 | 0.02 |
ENST00000234590.4
|
ENO1
|
enolase 1, (alpha) |
| chr2_-_99224915 | 0.02 |
ENST00000328709.3
ENST00000409997.1 |
COA5
|
cytochrome c oxidase assembly factor 5 |
| chr12_-_123215306 | 0.02 |
ENST00000356987.2
ENST00000436083.2 |
HCAR1
|
hydroxycarboxylic acid receptor 1 |
| chr4_+_184427235 | 0.02 |
ENST00000412117.1
ENST00000434682.2 |
ING2
|
inhibitor of growth family, member 2 |
| chrX_+_91034260 | 0.02 |
ENST00000395337.2
|
PCDH11X
|
protocadherin 11 X-linked |
| chr11_+_46402744 | 0.02 |
ENST00000533952.1
|
MDK
|
midkine (neurite growth-promoting factor 2) |
Network of associatons between targets according to the STRING database.
First level regulatory network of EP300
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0060356 | leucine import(GO:0060356) |
| 0.0 | 0.1 | GO:1903939 | negative regulation of histone H3-K9 dimethylation(GO:1900110) regulation of TORC2 signaling(GO:1903939) |
| 0.0 | 0.1 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 0.0 | 0.1 | GO:0002590 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 0.0 | 0.0 | GO:0045799 | positive regulation of chromatin assembly or disassembly(GO:0045799) |
| 0.0 | 0.0 | GO:0035623 | renal glucose absorption(GO:0035623) |
| 0.0 | 0.1 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.0 | 0.0 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.0 | 0.0 | GO:0072737 | response to diamide(GO:0072737) cellular response to diamide(GO:0072738) |
| 0.0 | 0.1 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.1 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) basic amino acid transmembrane transport(GO:1990822) |
| 0.0 | 0.0 | GO:0014054 | positive regulation of gamma-aminobutyric acid secretion(GO:0014054) |
| 0.0 | 0.1 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.1 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.0 | 0.0 | GO:0007506 | gonadal mesoderm development(GO:0007506) |
| 0.0 | 0.0 | GO:0002514 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 0.0 | 0.0 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 0.0 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.1 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.0 | 0.0 | GO:0052856 | NADHX epimerase activity(GO:0052856) NADPHX epimerase activity(GO:0052857) |
| 0.0 | 0.0 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.0 | 0.1 | GO:0001512 | dihydronicotinamide riboside quinone reductase activity(GO:0001512) melatonin binding(GO:1904408) |
| 0.0 | 0.1 | GO:0008158 | hedgehog receptor activity(GO:0008158) |