Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
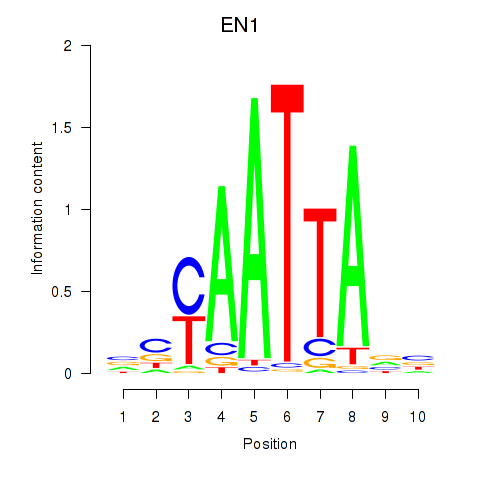
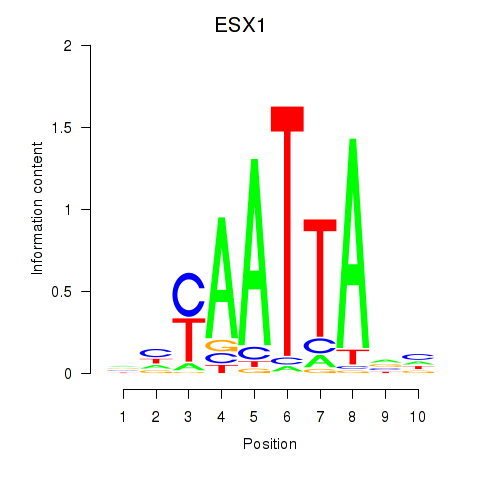
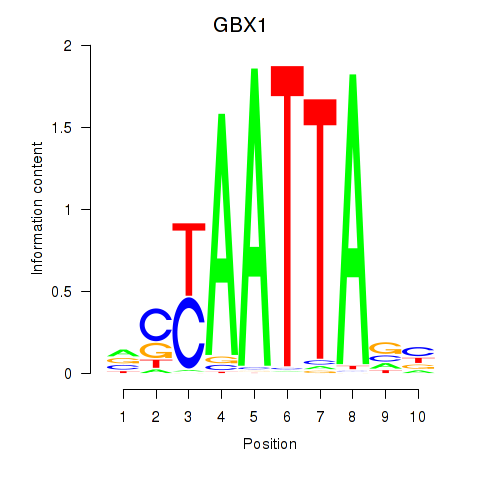
Results for EN1_ESX1_GBX1
Z-value: 0.42
Transcription factors associated with EN1_ESX1_GBX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EN1
|
ENSG00000163064.6 | engrailed homeobox 1 |
|
ESX1
|
ENSG00000123576.5 | ESX homeobox 1 |
|
GBX1
|
ENSG00000164900.4 | gastrulation brain homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EN1 | hg19_v2_chr2_-_119605253_119605264 | 0.69 | 3.1e-01 | Click! |
| GBX1 | hg19_v2_chr7_-_150864635_150864785 | -0.43 | 5.7e-01 | Click! |
Activity profile of EN1_ESX1_GBX1 motif
Sorted Z-values of EN1_ESX1_GBX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_105416039 | 0.51 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr16_+_53133070 | 0.49 |
ENST00000565832.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr17_-_64225508 | 0.48 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr15_-_37393406 | 0.37 |
ENST00000338564.5
ENST00000558313.1 ENST00000340545.5 |
MEIS2
|
Meis homeobox 2 |
| chr10_-_101825151 | 0.35 |
ENST00000441382.1
|
CPN1
|
carboxypeptidase N, polypeptide 1 |
| chr10_-_93392811 | 0.33 |
ENST00000238994.5
|
PPP1R3C
|
protein phosphatase 1, regulatory subunit 3C |
| chr11_+_77532233 | 0.29 |
ENST00000525409.1
|
AAMDC
|
adipogenesis associated, Mth938 domain containing |
| chr17_+_59489112 | 0.28 |
ENST00000335108.2
|
C17orf82
|
chromosome 17 open reading frame 82 |
| chr6_+_43968306 | 0.28 |
ENST00000442114.2
ENST00000336600.5 ENST00000439969.2 |
C6orf223
|
chromosome 6 open reading frame 223 |
| chr3_-_112360116 | 0.28 |
ENST00000206423.3
ENST00000439685.2 |
CCDC80
|
coiled-coil domain containing 80 |
| chr12_-_10978957 | 0.25 |
ENST00000240619.2
|
TAS2R10
|
taste receptor, type 2, member 10 |
| chr16_+_12059091 | 0.25 |
ENST00000562385.1
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr20_-_45976816 | 0.24 |
ENST00000441977.1
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr2_-_73460334 | 0.24 |
ENST00000258083.2
|
PRADC1
|
protease-associated domain containing 1 |
| chr5_+_31193847 | 0.24 |
ENST00000514738.1
ENST00000265071.2 |
CDH6
|
cadherin 6, type 2, K-cadherin (fetal kidney) |
| chr7_+_99717230 | 0.20 |
ENST00000262932.3
|
CNPY4
|
canopy FGF signaling regulator 4 |
| chr14_-_104181771 | 0.20 |
ENST00000554913.1
ENST00000554974.1 ENST00000553361.1 ENST00000555055.1 ENST00000555964.1 ENST00000556682.1 ENST00000445556.1 ENST00000553332.1 ENST00000352127.7 |
XRCC3
|
X-ray repair complementing defective repair in Chinese hamster cells 3 |
| chr1_-_234667504 | 0.20 |
ENST00000421207.1
ENST00000435574.1 |
RP5-855F14.1
|
RP5-855F14.1 |
| chr19_-_47137942 | 0.20 |
ENST00000300873.4
|
GNG8
|
guanine nucleotide binding protein (G protein), gamma 8 |
| chr8_+_98881268 | 0.19 |
ENST00000254898.5
ENST00000524308.1 ENST00000522025.2 |
MATN2
|
matrilin 2 |
| chr14_-_51027838 | 0.18 |
ENST00000555216.1
|
MAP4K5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr2_-_55237484 | 0.18 |
ENST00000394609.2
|
RTN4
|
reticulon 4 |
| chr14_+_104182105 | 0.18 |
ENST00000311141.2
|
ZFYVE21
|
zinc finger, FYVE domain containing 21 |
| chr9_-_3469181 | 0.18 |
ENST00000366116.2
|
AL365202.1
|
Uncharacterized protein |
| chr19_+_45973120 | 0.18 |
ENST00000592811.1
ENST00000586615.1 |
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B |
| chr11_-_31531121 | 0.18 |
ENST00000532287.1
ENST00000526776.1 ENST00000534812.1 ENST00000529749.1 ENST00000278200.1 ENST00000530023.1 ENST00000533642.1 |
IMMP1L
|
IMP1 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr1_+_84767289 | 0.17 |
ENST00000394834.3
ENST00000370669.1 |
SAMD13
|
sterile alpha motif domain containing 13 |
| chr6_-_26032288 | 0.17 |
ENST00000244661.2
|
HIST1H3B
|
histone cluster 1, H3b |
| chr8_+_98900132 | 0.17 |
ENST00000520016.1
|
MATN2
|
matrilin 2 |
| chr7_+_138145076 | 0.17 |
ENST00000343526.4
|
TRIM24
|
tripartite motif containing 24 |
| chr15_+_89164520 | 0.17 |
ENST00000332810.3
|
AEN
|
apoptosis enhancing nuclease |
| chr11_-_111794446 | 0.17 |
ENST00000527950.1
|
CRYAB
|
crystallin, alpha B |
| chr5_+_148737562 | 0.17 |
ENST00000274569.4
|
PCYOX1L
|
prenylcysteine oxidase 1 like |
| chr2_+_158114051 | 0.17 |
ENST00000259056.4
|
GALNT5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) |
| chrX_-_71458802 | 0.16 |
ENST00000373657.1
ENST00000334463.3 |
ERCC6L
|
excision repair cross-complementing rodent repair deficiency, complementation group 6-like |
| chr13_-_67802549 | 0.16 |
ENST00000328454.5
ENST00000377865.2 |
PCDH9
|
protocadherin 9 |
| chr6_+_26087509 | 0.16 |
ENST00000397022.3
ENST00000353147.5 ENST00000352392.4 ENST00000349999.4 ENST00000317896.7 ENST00000357618.5 ENST00000470149.1 ENST00000336625.8 ENST00000461397.1 ENST00000488199.1 |
HFE
|
hemochromatosis |
| chr7_+_77428066 | 0.15 |
ENST00000422959.2
ENST00000307305.8 ENST00000424760.1 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr1_-_21625486 | 0.15 |
ENST00000481130.2
|
ECE1
|
endothelin converting enzyme 1 |
| chr9_-_116840728 | 0.15 |
ENST00000265132.3
|
AMBP
|
alpha-1-microglobulin/bikunin precursor |
| chr13_-_96329048 | 0.15 |
ENST00000606011.1
ENST00000499499.2 |
DNAJC3-AS1
|
DNAJC3 antisense RNA 1 (head to head) |
| chr12_-_28122980 | 0.15 |
ENST00000395868.3
ENST00000534890.1 |
PTHLH
|
parathyroid hormone-like hormone |
| chr4_+_147096837 | 0.15 |
ENST00000296581.5
ENST00000502781.1 |
LSM6
|
LSM6 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr3_+_149191723 | 0.14 |
ENST00000305354.4
|
TM4SF4
|
transmembrane 4 L six family member 4 |
| chr16_+_69345243 | 0.14 |
ENST00000254950.11
|
VPS4A
|
vacuolar protein sorting 4 homolog A (S. cerevisiae) |
| chr19_-_52307357 | 0.14 |
ENST00000594900.1
|
FPR1
|
formyl peptide receptor 1 |
| chr4_-_89442940 | 0.14 |
ENST00000527353.1
|
PIGY
|
phosphatidylinositol glycan anchor biosynthesis, class Y |
| chr5_-_159827033 | 0.14 |
ENST00000523213.1
|
C5orf54
|
chromosome 5 open reading frame 54 |
| chr11_+_77532155 | 0.14 |
ENST00000532481.1
ENST00000526415.1 ENST00000393427.2 ENST00000527134.1 ENST00000304716.8 |
AAMDC
|
adipogenesis associated, Mth938 domain containing |
| chr6_-_111927449 | 0.13 |
ENST00000368761.5
ENST00000392556.4 ENST00000340026.6 |
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr9_-_5339873 | 0.13 |
ENST00000223862.1
ENST00000223858.4 |
RLN1
|
relaxin 1 |
| chr13_+_110958124 | 0.13 |
ENST00000400163.2
|
COL4A2
|
collagen, type IV, alpha 2 |
| chr14_-_55658323 | 0.13 |
ENST00000554067.1
ENST00000247191.2 |
DLGAP5
|
discs, large (Drosophila) homolog-associated protein 5 |
| chr6_-_109702885 | 0.13 |
ENST00000504373.1
|
CD164
|
CD164 molecule, sialomucin |
| chr8_-_42698292 | 0.13 |
ENST00000529779.1
|
THAP1
|
THAP domain containing, apoptosis associated protein 1 |
| chr19_+_50016610 | 0.13 |
ENST00000596975.1
|
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr1_+_62901968 | 0.13 |
ENST00000452143.1
ENST00000442679.1 ENST00000371146.1 |
USP1
|
ubiquitin specific peptidase 1 |
| chr12_-_56693758 | 0.13 |
ENST00000547298.1
ENST00000551936.1 ENST00000551253.1 ENST00000551473.1 |
CS
|
citrate synthase |
| chr1_-_197115818 | 0.13 |
ENST00000367409.4
ENST00000294732.7 |
ASPM
|
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
| chr15_+_59910132 | 0.13 |
ENST00000559200.1
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chr4_+_119200215 | 0.12 |
ENST00000602573.1
|
SNHG8
|
small nucleolar RNA host gene 8 (non-protein coding) |
| chr6_-_111927062 | 0.12 |
ENST00000359831.4
|
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr12_+_28410128 | 0.12 |
ENST00000381259.1
ENST00000381256.1 |
CCDC91
|
coiled-coil domain containing 91 |
| chr3_-_178103144 | 0.12 |
ENST00000417383.1
ENST00000418585.1 ENST00000411727.1 ENST00000439810.1 |
RP11-33A14.1
|
RP11-33A14.1 |
| chr5_+_150639360 | 0.12 |
ENST00000523004.1
|
GM2A
|
GM2 ganglioside activator |
| chr21_+_17909594 | 0.12 |
ENST00000441820.1
ENST00000602280.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr5_+_136070614 | 0.12 |
ENST00000502421.1
|
CTB-1I21.1
|
CTB-1I21.1 |
| chr1_+_117544366 | 0.12 |
ENST00000256652.4
ENST00000369470.1 |
CD101
|
CD101 molecule |
| chr19_+_50016411 | 0.12 |
ENST00000426395.3
ENST00000600273.1 ENST00000599988.1 |
FCGRT
|
Fc fragment of IgG, receptor, transporter, alpha |
| chr11_+_34663913 | 0.12 |
ENST00000532302.1
|
EHF
|
ets homologous factor |
| chr15_-_72565340 | 0.12 |
ENST00000568360.1
|
PARP6
|
poly (ADP-ribose) polymerase family, member 6 |
| chr6_+_26365443 | 0.12 |
ENST00000527422.1
ENST00000356386.2 ENST00000396934.3 ENST00000377708.2 ENST00000396948.1 ENST00000508906.2 |
BTN3A2
|
butyrophilin, subfamily 3, member A2 |
| chrX_-_100129128 | 0.12 |
ENST00000372960.4
ENST00000372964.1 ENST00000217885.5 |
NOX1
|
NADPH oxidase 1 |
| chr14_+_104182061 | 0.12 |
ENST00000216602.6
|
ZFYVE21
|
zinc finger, FYVE domain containing 21 |
| chr17_-_57229155 | 0.12 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr14_+_74551650 | 0.11 |
ENST00000554938.1
|
LIN52
|
lin-52 homolog (C. elegans) |
| chr1_-_151762900 | 0.11 |
ENST00000440583.2
|
TDRKH
|
tudor and KH domain containing |
| chr12_-_88974236 | 0.11 |
ENST00000228280.5
ENST00000552044.1 ENST00000357116.4 |
KITLG
|
KIT ligand |
| chr18_+_46065393 | 0.11 |
ENST00000256413.3
|
CTIF
|
CBP80/20-dependent translation initiation factor |
| chr8_+_22424551 | 0.11 |
ENST00000523348.1
|
SORBS3
|
sorbin and SH3 domain containing 3 |
| chr15_-_64673630 | 0.11 |
ENST00000558008.1
ENST00000559519.1 ENST00000380258.2 |
KIAA0101
|
KIAA0101 |
| chr11_-_71823266 | 0.11 |
ENST00000538919.1
ENST00000539395.1 ENST00000542531.1 |
ANAPC15
|
anaphase promoting complex subunit 15 |
| chr5_+_115177178 | 0.11 |
ENST00000316788.7
|
AP3S1
|
adaptor-related protein complex 3, sigma 1 subunit |
| chrX_+_77166172 | 0.11 |
ENST00000343533.5
ENST00000350425.4 ENST00000341514.6 |
ATP7A
|
ATPase, Cu++ transporting, alpha polypeptide |
| chr16_+_53412368 | 0.11 |
ENST00000565189.1
|
RP11-44F14.2
|
RP11-44F14.2 |
| chr12_+_7013897 | 0.11 |
ENST00000007969.8
ENST00000323702.5 |
LRRC23
|
leucine rich repeat containing 23 |
| chr7_+_77428149 | 0.11 |
ENST00000415251.2
ENST00000275575.7 |
PHTF2
|
putative homeodomain transcription factor 2 |
| chr15_+_58702742 | 0.11 |
ENST00000356113.6
ENST00000414170.3 |
LIPC
|
lipase, hepatic |
| chr9_+_2159850 | 0.11 |
ENST00000416751.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr1_+_28261492 | 0.11 |
ENST00000373894.3
|
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr8_-_102803163 | 0.10 |
ENST00000523645.1
ENST00000520346.1 ENST00000220931.6 ENST00000522448.1 ENST00000522951.1 ENST00000522252.1 ENST00000519098.1 |
NCALD
|
neurocalcin delta |
| chr14_+_32798547 | 0.10 |
ENST00000557354.1
ENST00000557102.1 ENST00000557272.1 |
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr9_-_112970436 | 0.10 |
ENST00000400613.4
|
C9orf152
|
chromosome 9 open reading frame 152 |
| chr7_-_72971934 | 0.10 |
ENST00000411832.1
|
BCL7B
|
B-cell CLL/lymphoma 7B |
| chr1_+_196912902 | 0.10 |
ENST00000476712.2
ENST00000367415.5 |
CFHR2
|
complement factor H-related 2 |
| chr2_+_113299990 | 0.10 |
ENST00000537335.1
ENST00000417433.2 |
POLR1B
|
polymerase (RNA) I polypeptide B, 128kDa |
| chr9_-_5304432 | 0.10 |
ENST00000416837.1
ENST00000308420.3 |
RLN2
|
relaxin 2 |
| chr1_+_28261533 | 0.10 |
ENST00000411604.1
ENST00000373888.4 |
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr8_+_27238147 | 0.10 |
ENST00000412793.1
|
PTK2B
|
protein tyrosine kinase 2 beta |
| chr12_-_28123206 | 0.10 |
ENST00000542963.1
ENST00000535992.1 |
PTHLH
|
parathyroid hormone-like hormone |
| chr15_-_64673665 | 0.10 |
ENST00000300035.4
|
KIAA0101
|
KIAA0101 |
| chr14_-_23426270 | 0.10 |
ENST00000557591.1
ENST00000397409.4 ENST00000490506.1 ENST00000554406.1 |
HAUS4
|
HAUS augmin-like complex, subunit 4 |
| chr14_+_39944025 | 0.10 |
ENST00000554328.1
ENST00000556620.1 ENST00000557197.1 |
RP11-111A21.1
|
RP11-111A21.1 |
| chr12_-_56694083 | 0.10 |
ENST00000552688.1
ENST00000548041.1 ENST00000551137.1 ENST00000551968.1 ENST00000542324.2 ENST00000546930.1 ENST00000549221.1 ENST00000550159.1 ENST00000550734.1 |
CS
|
citrate synthase |
| chr5_+_137673200 | 0.10 |
ENST00000434981.2
|
FAM53C
|
family with sequence similarity 53, member C |
| chr14_-_55658252 | 0.10 |
ENST00000395425.2
|
DLGAP5
|
discs, large (Drosophila) homolog-associated protein 5 |
| chr20_-_50722183 | 0.10 |
ENST00000371523.4
|
ZFP64
|
ZFP64 zinc finger protein |
| chr1_-_211848899 | 0.10 |
ENST00000366998.3
ENST00000540251.1 ENST00000366999.4 |
NEK2
|
NIMA-related kinase 2 |
| chr5_+_172068232 | 0.09 |
ENST00000520919.1
ENST00000522853.1 ENST00000369800.5 |
NEURL1B
|
neuralized E3 ubiquitin protein ligase 1B |
| chr20_-_50418972 | 0.09 |
ENST00000395997.3
|
SALL4
|
spalt-like transcription factor 4 |
| chr10_+_94451574 | 0.09 |
ENST00000492654.2
|
HHEX
|
hematopoietically expressed homeobox |
| chr13_-_36788718 | 0.09 |
ENST00000317764.6
ENST00000379881.3 |
SOHLH2
|
spermatogenesis and oogenesis specific basic helix-loop-helix 2 |
| chr9_-_77643307 | 0.09 |
ENST00000376834.3
ENST00000376830.3 |
C9orf41
|
chromosome 9 open reading frame 41 |
| chr16_+_103816 | 0.09 |
ENST00000383018.3
ENST00000417493.1 |
SNRNP25
|
small nuclear ribonucleoprotein 25kDa (U11/U12) |
| chr14_-_38064198 | 0.09 |
ENST00000250448.2
|
FOXA1
|
forkhead box A1 |
| chr1_+_28261621 | 0.09 |
ENST00000549094.1
|
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr8_+_107738240 | 0.09 |
ENST00000449762.2
ENST00000297447.6 |
OXR1
|
oxidation resistance 1 |
| chr17_+_43238438 | 0.09 |
ENST00000593138.1
ENST00000586681.1 |
HEXIM2
|
hexamethylene bis-acetamide inducible 2 |
| chr6_-_32908792 | 0.09 |
ENST00000418107.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr11_+_115498761 | 0.09 |
ENST00000424313.2
|
AP000997.1
|
AP000997.1 |
| chrX_-_100129320 | 0.09 |
ENST00000372966.3
|
NOX1
|
NADPH oxidase 1 |
| chr1_-_201140673 | 0.09 |
ENST00000367333.2
|
TMEM9
|
transmembrane protein 9 |
| chr12_+_56473939 | 0.09 |
ENST00000450146.2
|
ERBB3
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chrX_+_72783026 | 0.09 |
ENST00000373504.6
ENST00000373502.5 |
CHIC1
|
cysteine-rich hydrophobic domain 1 |
| chr4_+_119199904 | 0.09 |
ENST00000602483.1
ENST00000602819.1 |
SNHG8
|
small nucleolar RNA host gene 8 (non-protein coding) |
| chr13_-_46679144 | 0.09 |
ENST00000181383.4
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr3_+_151591422 | 0.09 |
ENST00000362032.5
|
SUCNR1
|
succinate receptor 1 |
| chr12_+_56473910 | 0.09 |
ENST00000411731.2
|
ERBB3
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr5_+_162864575 | 0.09 |
ENST00000512163.1
ENST00000393929.1 ENST00000340828.2 ENST00000511683.2 ENST00000510097.1 ENST00000511490.2 ENST00000510664.1 |
CCNG1
|
cyclin G1 |
| chr5_+_52776228 | 0.09 |
ENST00000256759.3
|
FST
|
follistatin |
| chr14_-_83262540 | 0.09 |
ENST00000554451.1
|
RP11-11K13.1
|
RP11-11K13.1 |
| chr1_-_36916011 | 0.09 |
ENST00000356637.5
ENST00000354267.3 ENST00000235532.5 |
OSCP1
|
organic solute carrier partner 1 |
| chr14_-_23426322 | 0.08 |
ENST00000555367.1
|
HAUS4
|
HAUS augmin-like complex, subunit 4 |
| chr8_+_41386725 | 0.08 |
ENST00000276533.3
ENST00000520710.1 ENST00000518671.1 |
GINS4
|
GINS complex subunit 4 (Sld5 homolog) |
| chr4_+_3388057 | 0.08 |
ENST00000538395.1
|
RGS12
|
regulator of G-protein signaling 12 |
| chr12_-_56694142 | 0.08 |
ENST00000550655.1
ENST00000548567.1 ENST00000551430.2 ENST00000351328.3 |
CS
|
citrate synthase |
| chr4_+_174089904 | 0.08 |
ENST00000265000.4
|
GALNT7
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7) |
| chr2_-_224467093 | 0.08 |
ENST00000305409.2
|
SCG2
|
secretogranin II |
| chr17_+_47448102 | 0.08 |
ENST00000576461.1
|
RP11-81K2.1
|
Uncharacterized protein |
| chr4_+_25915896 | 0.08 |
ENST00000514384.1
|
SMIM20
|
small integral membrane protein 20 |
| chr14_-_23426337 | 0.08 |
ENST00000342454.8
ENST00000555986.1 ENST00000541587.1 ENST00000554516.1 ENST00000347758.2 ENST00000206474.7 ENST00000555040.1 |
HAUS4
|
HAUS augmin-like complex, subunit 4 |
| chr7_-_130080977 | 0.08 |
ENST00000223208.5
|
CEP41
|
centrosomal protein 41kDa |
| chr14_+_74815116 | 0.08 |
ENST00000256362.4
|
VRTN
|
vertebrae development associated |
| chr10_+_5005598 | 0.08 |
ENST00000442997.1
|
AKR1C1
|
aldo-keto reductase family 1, member C1 |
| chr1_+_43291220 | 0.08 |
ENST00000372514.3
|
ERMAP
|
erythroblast membrane-associated protein (Scianna blood group) |
| chr12_+_107712173 | 0.08 |
ENST00000280758.5
ENST00000420571.2 |
BTBD11
|
BTB (POZ) domain containing 11 |
| chr1_+_207277590 | 0.08 |
ENST00000367070.3
|
C4BPA
|
complement component 4 binding protein, alpha |
| chr11_-_117748138 | 0.08 |
ENST00000527717.1
|
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr10_-_33405600 | 0.08 |
ENST00000414308.1
|
RP11-342D11.3
|
RP11-342D11.3 |
| chr1_+_62439037 | 0.08 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr5_+_81601166 | 0.08 |
ENST00000439350.1
|
ATP6AP1L
|
ATPase, H+ transporting, lysosomal accessory protein 1-like |
| chr22_+_42834029 | 0.08 |
ENST00000428765.1
|
CTA-126B4.7
|
CTA-126B4.7 |
| chr11_+_34664014 | 0.08 |
ENST00000527935.1
|
EHF
|
ets homologous factor |
| chr20_+_43990576 | 0.08 |
ENST00000372727.1
ENST00000414310.1 |
SYS1
|
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae) |
| chr18_+_32556892 | 0.08 |
ENST00000591734.1
ENST00000413393.1 ENST00000589180.1 ENST00000587359.1 |
MAPRE2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr9_+_116225999 | 0.08 |
ENST00000317613.6
|
RGS3
|
regulator of G-protein signaling 3 |
| chr12_-_57328187 | 0.08 |
ENST00000293502.1
|
SDR9C7
|
short chain dehydrogenase/reductase family 9C, member 7 |
| chr14_-_45252031 | 0.08 |
ENST00000556405.1
|
RP11-398E10.1
|
RP11-398E10.1 |
| chr6_-_711395 | 0.08 |
ENST00000606285.1
|
RP11-532F6.3
|
RP11-532F6.3 |
| chr12_-_121972556 | 0.07 |
ENST00000545022.1
|
KDM2B
|
lysine (K)-specific demethylase 2B |
| chr11_-_117747434 | 0.07 |
ENST00000529335.2
ENST00000530956.1 ENST00000260282.4 |
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr17_-_28661065 | 0.07 |
ENST00000328886.4
ENST00000538566.2 |
TMIGD1
|
transmembrane and immunoglobulin domain containing 1 |
| chr9_-_136933134 | 0.07 |
ENST00000303407.7
|
BRD3
|
bromodomain containing 3 |
| chr6_-_72130472 | 0.07 |
ENST00000426635.2
|
LINC00472
|
long intergenic non-protein coding RNA 472 |
| chr16_+_58283814 | 0.07 |
ENST00000443128.2
ENST00000219299.4 |
CCDC113
|
coiled-coil domain containing 113 |
| chr4_-_19458597 | 0.07 |
ENST00000505347.1
|
RP11-3J1.1
|
RP11-3J1.1 |
| chr1_-_21620877 | 0.07 |
ENST00000527991.1
|
ECE1
|
endothelin converting enzyme 1 |
| chr13_+_78315295 | 0.07 |
ENST00000351546.3
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr3_+_130569429 | 0.07 |
ENST00000505330.1
ENST00000504381.1 ENST00000507488.2 ENST00000393221.4 |
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chr5_+_142286887 | 0.07 |
ENST00000451259.1
|
ARHGAP26
|
Rho GTPase activating protein 26 |
| chr2_+_38152462 | 0.07 |
ENST00000354545.2
|
RMDN2
|
regulator of microtubule dynamics 2 |
| chr12_+_52056548 | 0.07 |
ENST00000545061.1
ENST00000355133.3 |
SCN8A
|
sodium channel, voltage gated, type VIII, alpha subunit |
| chr4_+_66536248 | 0.07 |
ENST00000514260.1
ENST00000507117.1 |
RP11-807H7.1
|
RP11-807H7.1 |
| chr3_+_167453493 | 0.07 |
ENST00000295777.5
ENST00000472747.2 |
SERPINI1
|
serpin peptidase inhibitor, clade I (neuroserpin), member 1 |
| chr17_+_18086392 | 0.07 |
ENST00000541285.1
|
ALKBH5
|
alkB, alkylation repair homolog 5 (E. coli) |
| chr15_-_65903407 | 0.07 |
ENST00000395644.4
ENST00000567744.1 ENST00000568573.1 ENST00000562830.1 ENST00000569491.1 ENST00000561769.1 |
VWA9
|
von Willebrand factor A domain containing 9 |
| chr11_-_71823796 | 0.07 |
ENST00000545680.1
ENST00000543587.1 ENST00000538393.1 ENST00000535234.1 ENST00000227618.4 ENST00000535503.1 |
ANAPC15
|
anaphase promoting complex subunit 15 |
| chr1_+_153746683 | 0.07 |
ENST00000271857.2
|
SLC27A3
|
solute carrier family 27 (fatty acid transporter), member 3 |
| chr14_-_23426231 | 0.07 |
ENST00000556915.1
|
HAUS4
|
HAUS augmin-like complex, subunit 4 |
| chr4_-_46911223 | 0.07 |
ENST00000396533.1
|
COX7B2
|
cytochrome c oxidase subunit VIIb2 |
| chr2_+_128293323 | 0.07 |
ENST00000389524.4
ENST00000428314.1 |
MYO7B
|
myosin VIIB |
| chr10_+_135050908 | 0.07 |
ENST00000325980.9
|
VENTX
|
VENT homeobox |
| chr16_-_46655538 | 0.07 |
ENST00000303383.3
|
SHCBP1
|
SHC SH2-domain binding protein 1 |
| chr2_-_220117867 | 0.07 |
ENST00000456818.1
ENST00000447205.1 |
TUBA4A
|
tubulin, alpha 4a |
| chr3_-_180397256 | 0.07 |
ENST00000442201.2
|
CCDC39
|
coiled-coil domain containing 39 |
| chr1_+_196788887 | 0.07 |
ENST00000320493.5
ENST00000367424.4 ENST00000367421.3 |
CFHR1
CFHR2
|
complement factor H-related 1 complement factor H-related 2 |
| chr1_+_236958554 | 0.07 |
ENST00000366577.5
ENST00000418145.2 |
MTR
|
5-methyltetrahydrofolate-homocysteine methyltransferase |
| chr15_+_89164560 | 0.06 |
ENST00000379231.3
ENST00000559528.1 |
AEN
|
apoptosis enhancing nuclease |
| chr9_-_75653627 | 0.06 |
ENST00000446946.1
|
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1 |
| chr7_+_127292234 | 0.06 |
ENST00000354725.3
|
SND1
|
staphylococcal nuclease and tudor domain containing 1 |
| chr5_+_52776449 | 0.06 |
ENST00000396947.3
|
FST
|
follistatin |
| chr17_+_36508826 | 0.06 |
ENST00000580660.1
|
SOCS7
|
suppressor of cytokine signaling 7 |
| chr7_-_55620433 | 0.06 |
ENST00000418904.1
|
VOPP1
|
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chr13_-_46679185 | 0.06 |
ENST00000439329.3
|
CPB2
|
carboxypeptidase B2 (plasma) |
| chr6_-_32908765 | 0.06 |
ENST00000416244.2
|
HLA-DMB
|
major histocompatibility complex, class II, DM beta |
| chr15_+_75080883 | 0.06 |
ENST00000567571.1
|
CSK
|
c-src tyrosine kinase |
| chrX_+_154444643 | 0.06 |
ENST00000286428.5
|
VBP1
|
von Hippel-Lindau binding protein 1 |
| chr12_-_118628350 | 0.06 |
ENST00000537952.1
ENST00000537822.1 |
TAOK3
|
TAO kinase 3 |
| chr15_+_69857515 | 0.06 |
ENST00000559477.1
|
RP11-279F6.1
|
RP11-279F6.1 |
| chr4_+_129732419 | 0.06 |
ENST00000510308.1
|
PHF17
|
jade family PHD finger 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of EN1_ESX1_GBX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0030070 | insulin processing(GO:0030070) |
| 0.1 | 0.2 | GO:0002416 | IgG immunoglobulin transcytosis in epithelial cells mediated by FcRn immunoglobulin receptor(GO:0002416) |
| 0.1 | 0.5 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.1 | 0.2 | GO:0071140 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 0.1 | 0.2 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.1 | 0.3 | GO:0034756 | regulation of iron ion transport(GO:0034756) regulation of iron ion transmembrane transport(GO:0034759) |
| 0.1 | 0.2 | GO:2001190 | peptide antigen assembly with MHC class II protein complex(GO:0002503) positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.1 | 0.2 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.0 | 0.1 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 | 0.6 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.1 | GO:0033024 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.0 | 0.2 | GO:0010814 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.0 | 0.2 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.0 | 0.1 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.0 | 0.1 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) gall bladder development(GO:0061010) |
| 0.0 | 0.1 | GO:0060738 | epithelial-mesenchymal signaling involved in prostate gland development(GO:0060738) |
| 0.0 | 0.2 | GO:1902952 | positive regulation of dendritic spine maintenance(GO:1902952) |
| 0.0 | 0.2 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.2 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.0 | GO:0001812 | positive regulation of type I hypersensitivity(GO:0001812) |
| 0.0 | 0.3 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.2 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.1 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.1 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.1 | GO:2000537 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.0 | 0.1 | GO:2000870 | regulation of progesterone secretion(GO:2000870) |
| 0.0 | 0.0 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.0 | 0.2 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.0 | GO:0048241 | epinephrine transport(GO:0048241) |
| 0.0 | 0.1 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.0 | 0.1 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) |
| 0.0 | 0.1 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.0 | 0.2 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.2 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.0 | 0.0 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.0 | 0.0 | GO:1900175 | regulation of nodal signaling pathway involved in determination of left/right asymmetry(GO:1900145) regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900175) |
| 0.0 | 0.1 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 | 0.1 | GO:0035513 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.0 | GO:1903625 | negative regulation of DNA catabolic process(GO:1903625) |
| 0.0 | 0.2 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.1 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.0 | 0.1 | GO:0034372 | very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.0 | 0.1 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.1 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.0 | 0.0 | GO:0033685 | negative regulation of luteinizing hormone secretion(GO:0033685) |
| 0.0 | 0.0 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.1 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.0 | GO:1990418 | response to insulin-like growth factor stimulus(GO:1990418) |
| 0.0 | 0.1 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.1 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.1 | GO:0010700 | negative regulation of norepinephrine secretion(GO:0010700) |
| 0.0 | 0.0 | GO:0051866 | general adaptation syndrome(GO:0051866) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.2 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.2 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.0 | 0.2 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.1 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.0 | 0.2 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.5 | GO:0031089 | platelet dense granule lumen(GO:0031089) |
| 0.0 | 0.3 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.1 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.2 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.1 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.0 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.1 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.1 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.0 | 0.3 | GO:0005614 | interstitial matrix(GO:0005614) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.3 | GO:0004108 | citrate (Si)-synthase activity(GO:0004108) citrate synthase activity(GO:0036440) |
| 0.1 | 0.3 | GO:0000285 | 1-phosphatidylinositol-3-phosphate 5-kinase activity(GO:0000285) |
| 0.1 | 0.4 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.1 | 0.2 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.1 | GO:0035248 | alpha-1,4-N-acetylgalactosaminyltransferase activity(GO:0035248) |
| 0.0 | 0.1 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.0 | 0.0 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.2 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.2 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.1 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.3 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.3 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.1 | GO:0004803 | transposase activity(GO:0004803) |
| 0.0 | 0.2 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.0 | 0.1 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.2 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.0 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.0 | 0.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.1 | GO:0047718 | androsterone dehydrogenase activity(GO:0047023) indanol dehydrogenase activity(GO:0047718) |
| 0.0 | 0.0 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.0 | 0.1 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.0 | 0.0 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.1 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.2 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.1 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.0 | GO:0031779 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.0 | GO:0005277 | acetylcholine transmembrane transporter activity(GO:0005277) norepinephrine transmembrane transporter activity(GO:0005333) acetate ester transmembrane transporter activity(GO:1901375) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |