Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for EMX2
Z-value: 2.00
Transcription factors associated with EMX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EMX2
|
ENSG00000170370.10 | empty spiracles homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EMX2 | hg19_v2_chr10_+_119302508_119302559 | -0.70 | 3.0e-01 | Click! |
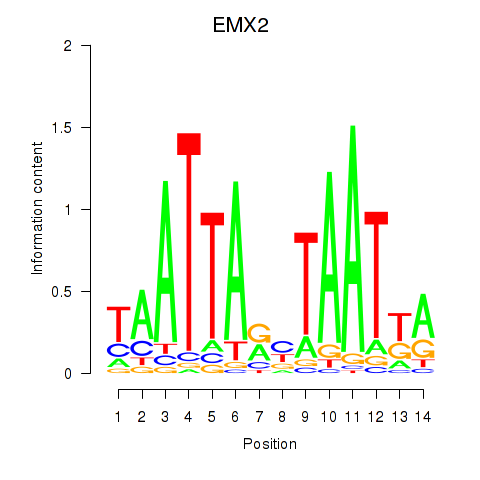
Activity profile of EMX2 motif
Sorted Z-values of EMX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_54466179 | 4.46 |
ENST00000270458.2
|
CACNG8
|
calcium channel, voltage-dependent, gamma subunit 8 |
| chr11_+_5710919 | 3.18 |
ENST00000379965.3
ENST00000425490.1 |
TRIM22
|
tripartite motif containing 22 |
| chr10_+_91092241 | 3.07 |
ENST00000371811.4
|
IFIT3
|
interferon-induced protein with tetratricopeptide repeats 3 |
| chr11_-_327537 | 2.76 |
ENST00000602735.1
|
IFITM3
|
interferon induced transmembrane protein 3 |
| chr1_+_79115503 | 2.49 |
ENST00000370747.4
ENST00000438486.1 ENST00000545124.1 |
IFI44
|
interferon-induced protein 44 |
| chr15_+_63188009 | 1.68 |
ENST00000557900.1
|
RP11-1069G10.2
|
RP11-1069G10.2 |
| chr11_-_104905840 | 1.64 |
ENST00000526568.1
ENST00000393136.4 ENST00000531166.1 ENST00000534497.1 ENST00000527979.1 ENST00000446369.1 ENST00000353247.5 ENST00000528974.1 ENST00000533400.1 ENST00000525825.1 ENST00000436863.3 |
CASP1
|
caspase 1, apoptosis-related cysteine peptidase |
| chr2_+_228736335 | 1.60 |
ENST00000440997.1
ENST00000545118.1 |
DAW1
|
dynein assembly factor with WDR repeat domains 1 |
| chr4_-_89205705 | 1.45 |
ENST00000295908.7
ENST00000510548.2 ENST00000508256.1 |
PPM1K
|
protein phosphatase, Mg2+/Mn2+ dependent, 1K |
| chr15_+_101417919 | 1.42 |
ENST00000561338.1
|
ALDH1A3
|
aldehyde dehydrogenase 1 family, member A3 |
| chr17_-_71223839 | 1.40 |
ENST00000579872.1
ENST00000580032.1 |
FAM104A
|
family with sequence similarity 104, member A |
| chr4_-_120243545 | 1.31 |
ENST00000274024.3
|
FABP2
|
fatty acid binding protein 2, intestinal |
| chrX_+_78003204 | 1.21 |
ENST00000435339.3
ENST00000514744.1 |
LPAR4
|
lysophosphatidic acid receptor 4 |
| chr11_-_18258342 | 1.19 |
ENST00000278222.4
|
SAA4
|
serum amyloid A4, constitutive |
| chr10_-_90611566 | 1.16 |
ENST00000371930.4
|
ANKRD22
|
ankyrin repeat domain 22 |
| chr22_+_50981079 | 1.13 |
ENST00000609268.1
|
CTA-384D8.34
|
CTA-384D8.34 |
| chr18_+_61637159 | 1.08 |
ENST00000397985.2
ENST00000353706.2 ENST00000542677.1 ENST00000397988.3 ENST00000448851.1 |
SERPINB8
|
serpin peptidase inhibitor, clade B (ovalbumin), member 8 |
| chr12_-_10007448 | 1.07 |
ENST00000538152.1
|
CLEC2B
|
C-type lectin domain family 2, member B |
| chr11_-_104916034 | 1.03 |
ENST00000528513.1
ENST00000375706.2 ENST00000375704.3 |
CARD16
|
caspase recruitment domain family, member 16 |
| chr8_-_87242589 | 0.89 |
ENST00000419776.2
ENST00000297524.3 |
SLC7A13
|
solute carrier family 7 (anionic amino acid transporter), member 13 |
| chr3_-_108248169 | 0.88 |
ENST00000273353.3
|
MYH15
|
myosin, heavy chain 15 |
| chr4_-_89205879 | 0.86 |
ENST00000608933.1
ENST00000315194.4 ENST00000514204.1 |
PPM1K
|
protein phosphatase, Mg2+/Mn2+ dependent, 1K |
| chr10_-_79397740 | 0.85 |
ENST00000372440.1
ENST00000480683.1 |
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr12_-_123187890 | 0.80 |
ENST00000328880.5
|
HCAR2
|
hydroxycarboxylic acid receptor 2 |
| chr11_-_104972158 | 0.79 |
ENST00000598974.1
ENST00000593315.1 ENST00000594519.1 ENST00000415981.2 ENST00000525374.1 ENST00000375707.1 |
CASP1
CARD16
CARD17
|
caspase 1, apoptosis-related cysteine peptidase caspase recruitment domain family, member 16 caspase recruitment domain family, member 17 |
| chr11_-_63376013 | 0.77 |
ENST00000540943.1
|
PLA2G16
|
phospholipase A2, group XVI |
| chr5_-_55412774 | 0.77 |
ENST00000434982.2
|
ANKRD55
|
ankyrin repeat domain 55 |
| chr3_-_48956818 | 0.76 |
ENST00000408959.2
|
ARIH2OS
|
ariadne homolog 2 opposite strand |
| chr15_-_78913628 | 0.74 |
ENST00000348639.3
|
CHRNA3
|
cholinergic receptor, nicotinic, alpha 3 (neuronal) |
| chr9_-_21368075 | 0.74 |
ENST00000449498.1
|
IFNA13
|
interferon, alpha 13 |
| chr1_-_92952433 | 0.72 |
ENST00000294702.5
|
GFI1
|
growth factor independent 1 transcription repressor |
| chr15_-_59500973 | 0.68 |
ENST00000560749.1
|
MYO1E
|
myosin IE |
| chr11_-_74800799 | 0.68 |
ENST00000305159.3
|
OR2AT4
|
olfactory receptor, family 2, subfamily AT, member 4 |
| chr5_+_34757309 | 0.67 |
ENST00000397449.1
|
RAI14
|
retinoic acid induced 14 |
| chr4_-_74864386 | 0.66 |
ENST00000296027.4
|
CXCL5
|
chemokine (C-X-C motif) ligand 5 |
| chr12_+_28605426 | 0.66 |
ENST00000542801.1
|
CCDC91
|
coiled-coil domain containing 91 |
| chr8_-_40200903 | 0.65 |
ENST00000522486.1
|
CTA-392C11.1
|
CTA-392C11.1 |
| chr2_-_198540751 | 0.64 |
ENST00000429081.1
|
RFTN2
|
raftlin family member 2 |
| chr22_+_39966758 | 0.63 |
ENST00000407673.1
ENST00000401624.1 ENST00000404898.1 ENST00000402142.3 ENST00000336649.4 ENST00000400164.3 |
CACNA1I
|
calcium channel, voltage-dependent, T type, alpha 1I subunit |
| chr17_-_39093672 | 0.63 |
ENST00000209718.3
ENST00000436344.3 ENST00000485751.1 |
KRT23
|
keratin 23 (histone deacetylase inducible) |
| chr6_-_134639180 | 0.62 |
ENST00000367858.5
|
SGK1
|
serum/glucocorticoid regulated kinase 1 |
| chr8_+_119294456 | 0.61 |
ENST00000366457.2
|
AC023590.1
|
Uncharacterized protein |
| chr12_+_25055243 | 0.61 |
ENST00000599478.1
|
AC026310.1
|
Protein 101060047 |
| chr1_+_192778161 | 0.60 |
ENST00000235382.5
|
RGS2
|
regulator of G-protein signaling 2, 24kDa |
| chr6_+_34725181 | 0.60 |
ENST00000244520.5
|
SNRPC
|
small nuclear ribonucleoprotein polypeptide C |
| chrX_+_100224676 | 0.59 |
ENST00000450049.2
|
ARL13A
|
ADP-ribosylation factor-like 13A |
| chr5_-_58571935 | 0.59 |
ENST00000503258.1
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr2_-_207082748 | 0.58 |
ENST00000407325.2
ENST00000411719.1 |
GPR1
|
G protein-coupled receptor 1 |
| chr17_+_62461569 | 0.58 |
ENST00000603557.1
ENST00000605096.1 |
MILR1
|
mast cell immunoglobulin-like receptor 1 |
| chr15_-_41166414 | 0.57 |
ENST00000220507.4
|
RHOV
|
ras homolog family member V |
| chr6_+_27833034 | 0.56 |
ENST00000357320.2
|
HIST1H2AL
|
histone cluster 1, H2al |
| chr11_-_46113756 | 0.55 |
ENST00000531959.1
|
PHF21A
|
PHD finger protein 21A |
| chr8_-_40200877 | 0.55 |
ENST00000521030.1
|
CTA-392C11.1
|
CTA-392C11.1 |
| chr5_-_140013275 | 0.55 |
ENST00000512545.1
ENST00000302014.6 ENST00000401743.2 |
CD14
|
CD14 molecule |
| chr5_+_179135246 | 0.55 |
ENST00000508787.1
|
CANX
|
calnexin |
| chr12_+_93096619 | 0.53 |
ENST00000397833.3
|
C12orf74
|
chromosome 12 open reading frame 74 |
| chr5_-_140013224 | 0.53 |
ENST00000498971.2
|
CD14
|
CD14 molecule |
| chr12_+_1099675 | 0.52 |
ENST00000545318.2
|
ERC1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr5_-_94417186 | 0.52 |
ENST00000312216.8
ENST00000512425.1 |
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr2_+_122494676 | 0.52 |
ENST00000455432.1
|
TSN
|
translin |
| chr12_+_93096759 | 0.52 |
ENST00000544406.2
|
C12orf74
|
chromosome 12 open reading frame 74 |
| chr17_-_34503867 | 0.51 |
ENST00000589443.1
ENST00000454519.3 ENST00000586369.1 |
TBC1D3B
|
TBC1 domain family, member 3B |
| chr14_+_52164820 | 0.51 |
ENST00000554167.1
|
FRMD6
|
FERM domain containing 6 |
| chr12_-_58220078 | 0.50 |
ENST00000549039.1
|
CTDSP2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr14_+_31959154 | 0.50 |
ENST00000550005.1
|
NUBPL
|
nucleotide binding protein-like |
| chr2_-_99917639 | 0.49 |
ENST00000308528.4
|
LYG1
|
lysozyme G-like 1 |
| chr15_+_64680003 | 0.49 |
ENST00000261884.3
|
TRIP4
|
thyroid hormone receptor interactor 4 |
| chr1_-_35450897 | 0.49 |
ENST00000373337.3
|
ZMYM6NB
|
ZMYM6 neighbor |
| chr16_+_22524844 | 0.48 |
ENST00000538606.1
ENST00000424340.1 ENST00000517539.1 ENST00000528249.1 |
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr12_+_60083118 | 0.48 |
ENST00000261187.4
ENST00000543448.1 |
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr1_-_110283660 | 0.46 |
ENST00000361066.2
|
GSTM3
|
glutathione S-transferase mu 3 (brain) |
| chr12_-_22063787 | 0.46 |
ENST00000544039.1
|
ABCC9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr1_+_151253991 | 0.45 |
ENST00000443959.1
|
ZNF687
|
zinc finger protein 687 |
| chr1_-_190446759 | 0.45 |
ENST00000367462.3
|
BRINP3
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr2_+_39117010 | 0.45 |
ENST00000409978.1
|
ARHGEF33
|
Rho guanine nucleotide exchange factor (GEF) 33 |
| chr2_+_56179262 | 0.45 |
ENST00000606639.1
|
RP11-481J13.1
|
RP11-481J13.1 |
| chr2_+_32502952 | 0.44 |
ENST00000238831.4
|
YIPF4
|
Yip1 domain family, member 4 |
| chr15_-_34635314 | 0.44 |
ENST00000557912.1
ENST00000328848.4 |
NOP10
|
NOP10 ribonucleoprotein |
| chr1_+_36549676 | 0.44 |
ENST00000207457.3
|
TEKT2
|
tektin 2 (testicular) |
| chr20_-_45530365 | 0.44 |
ENST00000414085.1
|
RP11-323C15.2
|
RP11-323C15.2 |
| chrX_+_36254051 | 0.43 |
ENST00000378657.4
|
CXorf30
|
chromosome X open reading frame 30 |
| chr16_+_15489603 | 0.43 |
ENST00000568766.1
ENST00000287594.7 |
RP11-1021N1.1
MPV17L
|
Uncharacterized protein MPV17 mitochondrial membrane protein-like |
| chr10_-_21186144 | 0.43 |
ENST00000377119.1
|
NEBL
|
nebulette |
| chr8_-_74495065 | 0.43 |
ENST00000523533.1
|
STAU2
|
staufen double-stranded RNA binding protein 2 |
| chr3_+_38080691 | 0.43 |
ENST00000308059.6
ENST00000346219.3 ENST00000452631.2 |
DLEC1
|
deleted in lung and esophageal cancer 1 |
| chr4_-_72649763 | 0.43 |
ENST00000513476.1
|
GC
|
group-specific component (vitamin D binding protein) |
| chr4_+_74347400 | 0.42 |
ENST00000226355.3
|
AFM
|
afamin |
| chr14_+_45464658 | 0.42 |
ENST00000555874.1
|
FAM179B
|
family with sequence similarity 179, member B |
| chrX_+_47444613 | 0.41 |
ENST00000445623.1
|
TIMP1
|
TIMP metallopeptidase inhibitor 1 |
| chr11_-_128894053 | 0.41 |
ENST00000392657.3
|
ARHGAP32
|
Rho GTPase activating protein 32 |
| chr8_+_30244580 | 0.41 |
ENST00000523115.1
ENST00000519647.1 |
RBPMS
|
RNA binding protein with multiple splicing |
| chr3_-_109056419 | 0.41 |
ENST00000335658.6
|
DPPA4
|
developmental pluripotency associated 4 |
| chr5_-_173217931 | 0.41 |
ENST00000522731.1
|
CTB-43E15.4
|
CTB-43E15.4 |
| chr16_-_29415350 | 0.40 |
ENST00000524087.1
|
NPIPB11
|
nuclear pore complex interacting protein family, member B11 |
| chr16_-_21868978 | 0.40 |
ENST00000357370.5
ENST00000451409.1 ENST00000341400.7 ENST00000518761.4 |
NPIPB4
|
nuclear pore complex interacting protein family, member B4 |
| chr11_-_107729887 | 0.40 |
ENST00000525815.1
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr22_+_45725524 | 0.40 |
ENST00000405548.3
|
FAM118A
|
family with sequence similarity 118, member A |
| chr1_+_111773349 | 0.39 |
ENST00000533831.2
|
CHI3L2
|
chitinase 3-like 2 |
| chrM_+_10758 | 0.39 |
ENST00000361381.2
|
MT-ND4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr11_+_47293795 | 0.39 |
ENST00000422579.1
|
MADD
|
MAP-kinase activating death domain |
| chr1_+_225600404 | 0.39 |
ENST00000366845.2
|
AC092811.1
|
AC092811.1 |
| chr4_+_74702214 | 0.38 |
ENST00000226317.5
ENST00000515050.1 |
CXCL6
|
chemokine (C-X-C motif) ligand 6 |
| chrX_+_36053908 | 0.38 |
ENST00000378660.2
|
CHDC2
|
calponin homology domain containing 2 |
| chr2_+_234826016 | 0.37 |
ENST00000324695.4
ENST00000433712.2 |
TRPM8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr4_-_185694872 | 0.37 |
ENST00000505492.1
|
ACSL1
|
acyl-CoA synthetase long-chain family member 1 |
| chr7_-_73153178 | 0.37 |
ENST00000437775.2
ENST00000222800.3 |
ABHD11
|
abhydrolase domain containing 11 |
| chr5_+_115358088 | 0.37 |
ENST00000600981.3
|
CTD-2287O16.3
|
CTD-2287O16.3 |
| chr9_+_71986182 | 0.37 |
ENST00000303068.7
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr5_-_94417314 | 0.36 |
ENST00000505208.1
|
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr11_+_10476851 | 0.36 |
ENST00000396553.2
|
AMPD3
|
adenosine monophosphate deaminase 3 |
| chr18_+_616672 | 0.36 |
ENST00000338387.7
|
CLUL1
|
clusterin-like 1 (retinal) |
| chrX_+_77359726 | 0.35 |
ENST00000442431.1
|
PGK1
|
phosphoglycerate kinase 1 |
| chr10_+_15074190 | 0.35 |
ENST00000428897.1
ENST00000413672.1 |
OLAH
|
oleoyl-ACP hydrolase |
| chr1_+_26496362 | 0.35 |
ENST00000374266.5
ENST00000270812.5 |
ZNF593
|
zinc finger protein 593 |
| chr16_-_21436459 | 0.35 |
ENST00000448012.2
ENST00000504841.2 ENST00000419180.2 |
NPIPB3
|
nuclear pore complex interacting protein family, member B3 |
| chr1_-_139379 | 0.35 |
ENST00000423372.3
|
AL627309.1
|
Uncharacterized protein |
| chr20_+_36405665 | 0.35 |
ENST00000373469.1
|
CTNNBL1
|
catenin, beta like 1 |
| chr8_-_133687813 | 0.34 |
ENST00000250173.1
ENST00000519595.1 |
LRRC6
|
leucine rich repeat containing 6 |
| chr3_+_132316081 | 0.34 |
ENST00000249887.2
|
ACKR4
|
atypical chemokine receptor 4 |
| chr13_+_108921977 | 0.33 |
ENST00000430559.1
ENST00000375887.4 |
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b |
| chr16_+_74330673 | 0.33 |
ENST00000219313.4
ENST00000540379.1 ENST00000567958.1 ENST00000568615.2 |
PSMD7
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 7 |
| chr12_+_78359999 | 0.33 |
ENST00000550503.1
|
NAV3
|
neuron navigator 3 |
| chr20_-_1447547 | 0.32 |
ENST00000476071.1
|
NSFL1C
|
NSFL1 (p97) cofactor (p47) |
| chr1_+_235530675 | 0.32 |
ENST00000366601.3
ENST00000406207.1 ENST00000543662.1 |
TBCE
|
tubulin folding cofactor E |
| chr6_+_110501621 | 0.32 |
ENST00000368930.1
ENST00000307731.1 |
CDC40
|
cell division cycle 40 |
| chr3_+_37441062 | 0.31 |
ENST00000426078.1
|
C3orf35
|
chromosome 3 open reading frame 35 |
| chr12_+_59989918 | 0.31 |
ENST00000547379.1
ENST00000549465.1 |
SLC16A7
|
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr3_-_52719546 | 0.31 |
ENST00000439181.1
ENST00000449505.1 |
PBRM1
|
polybromo 1 |
| chr6_+_34725263 | 0.30 |
ENST00000374018.1
ENST00000374017.3 |
SNRPC
|
small nuclear ribonucleoprotein polypeptide C |
| chr17_+_42422662 | 0.30 |
ENST00000593167.1
ENST00000585512.1 ENST00000591740.1 ENST00000592783.1 ENST00000587387.1 ENST00000588237.1 ENST00000589265.1 |
GRN
|
granulin |
| chr6_-_112081113 | 0.30 |
ENST00000517419.1
|
FYN
|
FYN oncogene related to SRC, FGR, YES |
| chr5_-_94417339 | 0.30 |
ENST00000429576.2
ENST00000508509.1 ENST00000510732.1 |
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr4_+_41540160 | 0.30 |
ENST00000503057.1
ENST00000511496.1 |
LIMCH1
|
LIM and calponin homology domains 1 |
| chr17_+_5031687 | 0.30 |
ENST00000250066.6
ENST00000304328.5 |
USP6
|
ubiquitin specific peptidase 6 (Tre-2 oncogene) |
| chrX_+_77359671 | 0.29 |
ENST00000373316.4
|
PGK1
|
phosphoglycerate kinase 1 |
| chr20_-_1447467 | 0.29 |
ENST00000353088.2
ENST00000350991.4 |
NSFL1C
|
NSFL1 (p97) cofactor (p47) |
| chr5_-_140013241 | 0.29 |
ENST00000519715.1
|
CD14
|
CD14 molecule |
| chr2_+_105050794 | 0.29 |
ENST00000429464.1
ENST00000414442.1 ENST00000447380.1 |
AC013402.2
|
long intergenic non-protein coding RNA 1102 |
| chr3_+_140981456 | 0.29 |
ENST00000504264.1
|
ACPL2
|
acid phosphatase-like 2 |
| chr4_-_168155169 | 0.29 |
ENST00000534949.1
ENST00000535728.1 |
SPOCK3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr11_-_107729287 | 0.29 |
ENST00000375682.4
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr16_+_28648975 | 0.28 |
ENST00000529716.1
|
NPIPB8
|
nuclear pore complex interacting protein family, member B8 |
| chr17_-_34757039 | 0.28 |
ENST00000455054.2
ENST00000308078.7 |
TBC1D3H
TBC1D3C
|
TBC1 domain family, member 3H TBC1 domain family, member 3C |
| chr1_+_234765057 | 0.28 |
ENST00000429269.1
|
LINC00184
|
long intergenic non-protein coding RNA 184 |
| chr10_-_79398250 | 0.28 |
ENST00000286627.5
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr3_-_151047327 | 0.28 |
ENST00000325602.5
|
P2RY13
|
purinergic receptor P2Y, G-protein coupled, 13 |
| chr2_-_39456673 | 0.28 |
ENST00000378803.1
ENST00000395035.3 |
CDKL4
|
cyclin-dependent kinase-like 4 |
| chr3_-_186288097 | 0.28 |
ENST00000446782.1
|
TBCCD1
|
TBCC domain containing 1 |
| chr16_+_28763108 | 0.28 |
ENST00000357796.3
ENST00000550983.1 |
NPIPB9
|
nuclear pore complex interacting protein family, member B9 |
| chr12_+_10366223 | 0.27 |
ENST00000545290.1
|
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr2_-_99871570 | 0.27 |
ENST00000333017.2
ENST00000409679.1 ENST00000423306.1 |
LYG2
|
lysozyme G-like 2 |
| chr4_+_56212505 | 0.27 |
ENST00000505210.1
|
SRD5A3
|
steroid 5 alpha-reductase 3 |
| chr19_-_43969796 | 0.27 |
ENST00000244333.3
|
LYPD3
|
LY6/PLAUR domain containing 3 |
| chr15_-_99057551 | 0.27 |
ENST00000558256.1
|
FAM169B
|
family with sequence similarity 169, member B |
| chr14_-_47351391 | 0.27 |
ENST00000399222.3
|
MDGA2
|
MAM domain containing glycosylphosphatidylinositol anchor 2 |
| chr17_+_42422637 | 0.27 |
ENST00000053867.3
ENST00000588143.1 |
GRN
|
granulin |
| chr16_-_28374829 | 0.27 |
ENST00000532254.1
|
NPIPB6
|
nuclear pore complex interacting protein family, member B6 |
| chr16_+_82068873 | 0.27 |
ENST00000566213.1
|
HSD17B2
|
hydroxysteroid (17-beta) dehydrogenase 2 |
| chr3_+_52245458 | 0.27 |
ENST00000459884.1
|
ALAS1
|
aminolevulinate, delta-, synthase 1 |
| chr4_+_26344754 | 0.26 |
ENST00000515573.1
|
RBPJ
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr16_-_29479154 | 0.25 |
ENST00000549950.1
|
RP11-345J4.3
|
Uncharacterized protein |
| chr1_+_56046710 | 0.25 |
ENST00000422374.1
|
RP11-466L17.1
|
RP11-466L17.1 |
| chr9_-_100684845 | 0.25 |
ENST00000375119.3
|
C9orf156
|
chromosome 9 open reading frame 156 |
| chr16_+_69599861 | 0.25 |
ENST00000354436.2
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive |
| chr11_-_26593649 | 0.25 |
ENST00000455601.2
|
MUC15
|
mucin 15, cell surface associated |
| chr3_-_20053741 | 0.25 |
ENST00000389050.4
|
PP2D1
|
protein phosphatase 2C-like domain containing 1 |
| chr1_-_212588157 | 0.25 |
ENST00000261455.4
ENST00000535273.1 |
TMEM206
|
transmembrane protein 206 |
| chr7_+_90338712 | 0.25 |
ENST00000265741.3
ENST00000406263.1 |
CDK14
|
cyclin-dependent kinase 14 |
| chr18_+_29769978 | 0.25 |
ENST00000269202.6
ENST00000581447.1 |
MEP1B
|
meprin A, beta |
| chr7_+_13141010 | 0.24 |
ENST00000443947.1
|
AC011288.2
|
AC011288.2 |
| chr3_+_11314099 | 0.24 |
ENST00000446450.2
ENST00000354956.5 ENST00000354449.3 ENST00000419112.1 |
ATG7
|
autophagy related 7 |
| chr4_-_122854612 | 0.24 |
ENST00000264811.5
|
TRPC3
|
transient receptor potential cation channel, subfamily C, member 3 |
| chr16_+_11038403 | 0.24 |
ENST00000409552.3
|
CLEC16A
|
C-type lectin domain family 16, member A |
| chr1_+_15668240 | 0.24 |
ENST00000444385.1
|
FHAD1
|
forkhead-associated (FHA) phosphopeptide binding domain 1 |
| chr3_-_37216055 | 0.24 |
ENST00000336686.4
|
LRRFIP2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr9_+_71944241 | 0.24 |
ENST00000257515.8
|
FAM189A2
|
family with sequence similarity 189, member A2 |
| chr18_+_21693306 | 0.24 |
ENST00000540918.2
|
TTC39C
|
tetratricopeptide repeat domain 39C |
| chr11_+_122709200 | 0.24 |
ENST00000227348.4
|
CRTAM
|
cytotoxic and regulatory T cell molecule |
| chr15_+_84115868 | 0.24 |
ENST00000427482.2
|
SH3GL3
|
SH3-domain GRB2-like 3 |
| chr4_+_56212270 | 0.23 |
ENST00000264228.4
|
SRD5A3
|
steroid 5 alpha-reductase 3 |
| chr13_-_30880979 | 0.23 |
ENST00000414289.1
|
KATNAL1
|
katanin p60 subunit A-like 1 |
| chr19_-_37697976 | 0.23 |
ENST00000588873.1
|
CTC-454I21.3
|
Uncharacterized protein; Zinc finger protein 585B |
| chr14_-_47812321 | 0.23 |
ENST00000357362.3
ENST00000486952.2 ENST00000426342.1 |
MDGA2
|
MAM domain containing glycosylphosphatidylinositol anchor 2 |
| chr12_+_101988774 | 0.23 |
ENST00000545503.2
ENST00000536007.1 ENST00000541119.1 ENST00000361466.2 ENST00000551300.1 ENST00000550270.1 |
MYBPC1
|
myosin binding protein C, slow type |
| chr2_+_74153953 | 0.23 |
ENST00000264093.4
ENST00000348222.1 |
DGUOK
|
deoxyguanosine kinase |
| chr3_+_113616317 | 0.23 |
ENST00000440446.2
ENST00000488680.1 |
GRAMD1C
|
GRAM domain containing 1C |
| chr16_+_11038345 | 0.23 |
ENST00000409790.1
|
CLEC16A
|
C-type lectin domain family 16, member A |
| chr1_+_244998918 | 0.23 |
ENST00000366528.3
|
COX20
|
COX20 cytochrome C oxidase assembly factor |
| chr7_-_101272576 | 0.23 |
ENST00000223167.4
|
MYL10
|
myosin, light chain 10, regulatory |
| chr19_-_41388657 | 0.22 |
ENST00000301146.4
ENST00000291764.3 |
CYP2A7
|
cytochrome P450, family 2, subfamily A, polypeptide 7 |
| chr6_-_76782371 | 0.22 |
ENST00000369950.3
ENST00000369963.3 |
IMPG1
|
interphotoreceptor matrix proteoglycan 1 |
| chrX_+_154254960 | 0.22 |
ENST00000369498.3
|
FUNDC2
|
FUN14 domain containing 2 |
| chr10_+_122610853 | 0.22 |
ENST00000604585.1
|
WDR11
|
WD repeat domain 11 |
| chr12_+_110011571 | 0.22 |
ENST00000539696.1
ENST00000228510.3 ENST00000392727.3 |
MVK
|
mevalonate kinase |
| chr12_-_53171128 | 0.22 |
ENST00000332411.2
|
KRT76
|
keratin 76 |
| chr10_-_79398127 | 0.22 |
ENST00000372443.1
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr1_+_174844645 | 0.22 |
ENST00000486220.1
|
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr4_+_158493642 | 0.22 |
ENST00000507108.1
ENST00000455598.1 ENST00000509450.1 |
RP11-364P22.1
|
RP11-364P22.1 |
| chr19_+_14693888 | 0.21 |
ENST00000547437.1
ENST00000397439.2 ENST00000417570.1 |
CLEC17A
|
C-type lectin domain family 17, member A |
| chr2_+_157330081 | 0.21 |
ENST00000409674.1
|
GPD2
|
glycerol-3-phosphate dehydrogenase 2 (mitochondrial) |
Network of associatons between targets according to the STRING database.
First level regulatory network of EMX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.4 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.5 | 1.4 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.2 | 0.7 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.2 | 3.2 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.2 | 1.4 | GO:0071725 | toll-like receptor TLR1:TLR2 signaling pathway(GO:0038123) response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.2 | 0.8 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.2 | 4.5 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 | 0.4 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.1 | 2.8 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.1 | 1.0 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.1 | 0.8 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.1 | 0.8 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.1 | 1.3 | GO:0034465 | response to carbon monoxide(GO:0034465) |
| 0.1 | 0.5 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.1 | 0.9 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.8 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 0.5 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.1 | 0.5 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.1 | 0.7 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.1 | 0.3 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.1 | 0.2 | GO:0039521 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) suppression by virus of host autophagy(GO:0039521) |
| 0.1 | 0.3 | GO:0048936 | peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.1 | 0.7 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.1 | 0.5 | GO:0070458 | establishment of blood-nerve barrier(GO:0008065) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.1 | 0.8 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 0.6 | GO:0071877 | relaxation of vascular smooth muscle(GO:0060087) regulation of adrenergic receptor signaling pathway(GO:0071877) |
| 0.1 | 0.2 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 0.2 | GO:0098507 | polynucleotide 5' dephosphorylation(GO:0098507) |
| 0.1 | 2.5 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.1 | 0.5 | GO:0003383 | apical constriction(GO:0003383) |
| 0.1 | 0.2 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.1 | 0.2 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.1 | 0.2 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.1 | 0.4 | GO:0050955 | thermoception(GO:0050955) |
| 0.1 | 0.6 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.1 | 0.4 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.1 | 0.9 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.4 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.6 | GO:0032596 | protein transport into membrane raft(GO:0032596) |
| 0.0 | 0.3 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.1 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.0 | 0.7 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.3 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.2 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.0 | 0.2 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.0 | 0.4 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.2 | GO:1901545 | cellular response to raffinose(GO:0097403) response to raffinose(GO:1901545) |
| 0.0 | 0.2 | GO:0035668 | TRAM-dependent toll-like receptor signaling pathway(GO:0035668) TRAM-dependent toll-like receptor 4 signaling pathway(GO:0035669) |
| 0.0 | 3.2 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.0 | 0.6 | GO:0033004 | negative regulation of mast cell activation(GO:0033004) |
| 0.0 | 1.0 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.0 | 0.5 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 0.2 | GO:0044026 | DNA hypermethylation(GO:0044026) |
| 0.0 | 0.2 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.0 | 0.5 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 0.2 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.2 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.0 | 1.1 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 1.2 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.1 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.0 | 0.4 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.4 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.2 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.4 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.1 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.0 | 0.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.4 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.0 | 0.5 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.2 | GO:0002857 | positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.1 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.3 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.0 | 0.6 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.0 | 0.3 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.2 | GO:0038129 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) ERBB3 signaling pathway(GO:0038129) |
| 0.0 | 0.4 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.5 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.1 | GO:0014004 | microglia differentiation(GO:0014004) microglia development(GO:0014005) cellular response to norepinephrine stimulus(GO:0071874) |
| 0.0 | 0.3 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 1.2 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.1 | GO:0042109 | lymphotoxin A production(GO:0032641) lymphotoxin A biosynthetic process(GO:0042109) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.0 | 0.2 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.1 | GO:0038042 | dimeric G-protein coupled receptor signaling pathway(GO:0038042) calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.0 | 0.3 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.0 | 0.2 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.0 | 0.2 | GO:0035898 | parathyroid hormone secretion(GO:0035898) post-embryonic body morphogenesis(GO:0040032) regulation of parathyroid hormone secretion(GO:2000828) |
| 0.0 | 0.4 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.0 | 0.6 | GO:0045956 | positive regulation of calcium ion-dependent exocytosis(GO:0045956) |
| 0.0 | 0.3 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.0 | 0.4 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.5 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.1 | GO:0002767 | immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 0.0 | 0.8 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.0 | 0.3 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.3 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.2 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.1 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.0 | 0.5 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 | 0.2 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.7 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.1 | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.0 | 0.4 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.0 | 0.1 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 0.3 | GO:0003298 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.2 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.2 | GO:0006527 | arginine catabolic process(GO:0006527) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.5 | GO:0097179 | protease inhibitor complex(GO:0097179) |
| 0.2 | 1.4 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 0.6 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.1 | 4.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 0.5 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 0.9 | GO:0000243 | commitment complex(GO:0000243) |
| 0.1 | 0.4 | GO:0090661 | box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.2 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.0 | 1.0 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.5 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.2 | GO:0035370 | UBC13-UEV1A complex(GO:0035370) |
| 0.0 | 3.2 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.8 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.2 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.5 | GO:0044233 | ER-mitochondrion membrane contact site(GO:0044233) |
| 0.0 | 0.2 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.2 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.2 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.3 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.5 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.2 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.1 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.3 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.5 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.0 | 1.1 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.3 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.2 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.4 | GO:0005685 | U1 snRNP(GO:0005685) |
| 0.0 | 0.1 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.0 | 0.5 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.6 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.5 | GO:0009986 | cell surface(GO:0009986) |
| 0.0 | 0.1 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.3 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 1.3 | GO:0031903 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.2 | 1.4 | GO:0016019 | peptidoglycan receptor activity(GO:0016019) |
| 0.2 | 0.6 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.2 | 0.6 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.2 | 0.8 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.2 | 3.5 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.2 | 0.8 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.1 | 1.3 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.1 | 0.4 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.1 | 0.5 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.1 | 0.4 | GO:0016297 | oleoyl-[acyl-carrier-protein] hydrolase activity(GO:0004320) myristoyl-[acyl-carrier-protein] hydrolase activity(GO:0016295) palmitoyl-[acyl-carrier-protein] hydrolase activity(GO:0016296) acyl-[acyl-carrier-protein] hydrolase activity(GO:0016297) |
| 0.1 | 0.3 | GO:0000035 | acyl binding(GO:0000035) phosphopantetheine binding(GO:0031177) |
| 0.1 | 0.8 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.1 | 1.2 | GO:0070915 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 0.4 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.1 | 1.4 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.1 | 0.3 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.1 | 4.5 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 0.8 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.1 | 0.2 | GO:0019779 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.1 | 1.0 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.2 | GO:0004651 | polynucleotide 5'-phosphatase activity(GO:0004651) |
| 0.1 | 0.3 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.1 | 0.4 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.1 | 0.3 | GO:0047006 | 17-alpha,20-alpha-dihydroxypregn-4-en-3-one dehydrogenase activity(GO:0047006) |
| 0.1 | 0.2 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.0 | 0.2 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.0 | 0.4 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.2 | GO:0042806 | fucose binding(GO:0042806) |
| 0.0 | 0.9 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.4 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.5 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.6 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.1 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.0 | 0.2 | GO:0052656 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.7 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.5 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
| 0.0 | 1.3 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.4 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.1 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.2 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.7 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 1.2 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.2 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.5 | GO:0043295 | glutathione binding(GO:0043295) |
| 0.0 | 0.2 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.4 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.5 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.7 | GO:0000146 | microfilament motor activity(GO:0000146) |
| 0.0 | 0.4 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.1 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.1 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.0 | 1.9 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.4 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.4 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.3 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.1 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.2 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 0.5 | GO:0004950 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.0 | 0.3 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.3 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.3 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.7 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.4 | GO:0032813 | death receptor binding(GO:0005123) tumor necrosis factor receptor superfamily binding(GO:0032813) |
| 0.0 | 0.7 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.2 | GO:0008392 | arachidonic acid monooxygenase activity(GO:0008391) arachidonic acid epoxygenase activity(GO:0008392) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.4 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 1.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.9 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.5 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.6 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.5 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 1.5 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 1.5 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 0.8 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.1 | 5.8 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 0.8 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 3.9 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.5 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.8 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.5 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.6 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.6 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 1.2 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.2 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.3 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.2 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.3 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |