Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
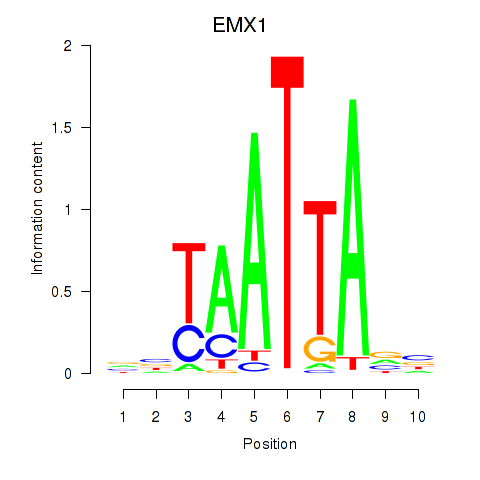
Results for EMX1
Z-value: 0.49
Transcription factors associated with EMX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
EMX1
|
ENSG00000135638.9 | empty spiracles homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| EMX1 | hg19_v2_chr2_+_73144604_73144654 | -0.94 | 5.6e-02 | Click! |
Activity profile of EMX1 motif
Sorted Z-values of EMX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_92777606 | 0.77 |
ENST00000437805.1
ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L
|
sterile alpha motif domain containing 9-like |
| chr17_+_62223320 | 0.40 |
ENST00000580828.1
ENST00000582965.1 |
SNORA76
|
small nucleolar RNA, H/ACA box 76 |
| chr14_-_37051798 | 0.30 |
ENST00000258829.5
|
NKX2-8
|
NK2 homeobox 8 |
| chr18_-_61329118 | 0.30 |
ENST00000332821.8
ENST00000283752.5 |
SERPINB3
|
serpin peptidase inhibitor, clade B (ovalbumin), member 3 |
| chr12_-_91546926 | 0.29 |
ENST00000550758.1
|
DCN
|
decorin |
| chr4_-_8873531 | 0.29 |
ENST00000400677.3
|
HMX1
|
H6 family homeobox 1 |
| chr11_-_33913708 | 0.24 |
ENST00000257818.2
|
LMO2
|
LIM domain only 2 (rhombotin-like 1) |
| chr18_-_59415987 | 0.24 |
ENST00000590199.1
ENST00000590968.1 |
RP11-879F14.1
|
RP11-879F14.1 |
| chr7_-_33102338 | 0.22 |
ENST00000610140.1
|
NT5C3A
|
5'-nucleotidase, cytosolic IIIA |
| chr12_+_1099675 | 0.22 |
ENST00000545318.2
|
ERC1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr3_-_108248169 | 0.21 |
ENST00000273353.3
|
MYH15
|
myosin, heavy chain 15 |
| chr11_-_111649074 | 0.21 |
ENST00000534218.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr3_-_143567262 | 0.21 |
ENST00000474151.1
ENST00000316549.6 |
SLC9A9
|
solute carrier family 9, subfamily A (NHE9, cation proton antiporter 9), member 9 |
| chr19_-_51522955 | 0.20 |
ENST00000358789.3
|
KLK10
|
kallikrein-related peptidase 10 |
| chrX_-_18690210 | 0.20 |
ENST00000379984.3
|
RS1
|
retinoschisin 1 |
| chr14_-_95236551 | 0.19 |
ENST00000238558.3
|
GSC
|
goosecoid homeobox |
| chr4_-_89205705 | 0.19 |
ENST00000295908.7
ENST00000510548.2 ENST00000508256.1 |
PPM1K
|
protein phosphatase, Mg2+/Mn2+ dependent, 1K |
| chr7_-_33102399 | 0.19 |
ENST00000242210.7
|
NT5C3A
|
5'-nucleotidase, cytosolic IIIA |
| chrX_+_100224676 | 0.19 |
ENST00000450049.2
|
ARL13A
|
ADP-ribosylation factor-like 13A |
| chr17_-_71223839 | 0.18 |
ENST00000579872.1
ENST00000580032.1 |
FAM104A
|
family with sequence similarity 104, member A |
| chr22_+_18632666 | 0.18 |
ENST00000215794.7
|
USP18
|
ubiquitin specific peptidase 18 |
| chr10_-_75351088 | 0.18 |
ENST00000451492.1
ENST00000413442.1 |
USP54
|
ubiquitin specific peptidase 54 |
| chr3_+_174158732 | 0.17 |
ENST00000434257.1
|
NAALADL2
|
N-acetylated alpha-linked acidic dipeptidase-like 2 |
| chr18_-_3220106 | 0.17 |
ENST00000356443.4
ENST00000400569.3 |
MYOM1
|
myomesin 1 |
| chr1_-_109399682 | 0.17 |
ENST00000369995.3
ENST00000370001.3 |
AKNAD1
|
AKNA domain containing 1 |
| chr17_+_79495397 | 0.17 |
ENST00000417245.2
ENST00000334850.7 |
FSCN2
|
fascin homolog 2, actin-bundling protein, retinal (Strongylocentrotus purpuratus) |
| chrM_+_8366 | 0.17 |
ENST00000361851.1
|
MT-ATP8
|
mitochondrially encoded ATP synthase 8 |
| chr14_-_21737551 | 0.16 |
ENST00000554891.1
ENST00000555883.1 ENST00000553753.1 ENST00000555914.1 ENST00000557336.1 ENST00000555215.1 ENST00000556628.1 ENST00000555137.1 ENST00000556226.1 ENST00000555309.1 ENST00000556142.1 ENST00000554969.1 ENST00000554455.1 ENST00000556513.1 ENST00000557201.1 ENST00000420743.2 ENST00000557768.1 ENST00000553300.1 ENST00000554383.1 ENST00000554539.1 |
HNRNPC
|
heterogeneous nuclear ribonucleoprotein C (C1/C2) |
| chrM_+_14741 | 0.16 |
ENST00000361789.2
|
MT-CYB
|
mitochondrially encoded cytochrome b |
| chrX_+_78003204 | 0.16 |
ENST00000435339.3
ENST00000514744.1 |
LPAR4
|
lysophosphatidic acid receptor 4 |
| chr12_+_19358192 | 0.16 |
ENST00000538305.1
|
PLEKHA5
|
pleckstrin homology domain containing, family A member 5 |
| chr9_-_117267717 | 0.16 |
ENST00000374057.3
|
DFNB31
|
deafness, autosomal recessive 31 |
| chr11_-_104905840 | 0.15 |
ENST00000526568.1
ENST00000393136.4 ENST00000531166.1 ENST00000534497.1 ENST00000527979.1 ENST00000446369.1 ENST00000353247.5 ENST00000528974.1 ENST00000533400.1 ENST00000525825.1 ENST00000436863.3 |
CASP1
|
caspase 1, apoptosis-related cysteine peptidase |
| chr12_+_12510352 | 0.15 |
ENST00000298571.6
|
LOH12CR1
|
loss of heterozygosity, 12, chromosomal region 1 |
| chr2_-_207082748 | 0.15 |
ENST00000407325.2
ENST00000411719.1 |
GPR1
|
G protein-coupled receptor 1 |
| chr6_-_109776901 | 0.15 |
ENST00000431946.1
|
MICAL1
|
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr15_+_80351910 | 0.15 |
ENST00000261749.6
ENST00000561060.1 |
ZFAND6
|
zinc finger, AN1-type domain 6 |
| chr7_+_100860949 | 0.14 |
ENST00000305105.2
|
ZNHIT1
|
zinc finger, HIT-type containing 1 |
| chr18_+_3447572 | 0.14 |
ENST00000548489.2
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr10_+_35484793 | 0.14 |
ENST00000488741.1
ENST00000474931.1 ENST00000468236.1 ENST00000344351.5 ENST00000490511.1 |
CREM
|
cAMP responsive element modulator |
| chr14_+_32798462 | 0.14 |
ENST00000280979.4
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr8_-_124553437 | 0.14 |
ENST00000517956.1
ENST00000443022.2 |
FBXO32
|
F-box protein 32 |
| chr17_+_19091325 | 0.13 |
ENST00000584923.1
|
SNORD3A
|
small nucleolar RNA, C/D box 3A |
| chr14_+_39944025 | 0.13 |
ENST00000554328.1
ENST00000556620.1 ENST00000557197.1 |
RP11-111A21.1
|
RP11-111A21.1 |
| chr20_+_60174827 | 0.13 |
ENST00000543233.1
|
CDH4
|
cadherin 4, type 1, R-cadherin (retinal) |
| chr6_+_31895254 | 0.13 |
ENST00000299367.5
ENST00000442278.2 |
C2
|
complement component 2 |
| chrM_+_8527 | 0.13 |
ENST00000361899.2
|
MT-ATP6
|
mitochondrially encoded ATP synthase 6 |
| chr19_+_4007644 | 0.13 |
ENST00000262971.2
|
PIAS4
|
protein inhibitor of activated STAT, 4 |
| chr8_-_49833978 | 0.12 |
ENST00000020945.1
|
SNAI2
|
snail family zinc finger 2 |
| chr19_+_11071546 | 0.12 |
ENST00000358026.2
|
SMARCA4
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| chr17_-_19015945 | 0.12 |
ENST00000573866.2
|
SNORD3D
|
small nucleolar RNA, C/D box 3D |
| chr15_+_62853562 | 0.12 |
ENST00000561311.1
|
TLN2
|
talin 2 |
| chr22_+_30752606 | 0.12 |
ENST00000399824.2
ENST00000405659.1 ENST00000338306.3 |
CCDC157
|
coiled-coil domain containing 157 |
| chr15_+_63188009 | 0.12 |
ENST00000557900.1
|
RP11-1069G10.2
|
RP11-1069G10.2 |
| chr12_-_112123524 | 0.12 |
ENST00000327551.6
|
BRAP
|
BRCA1 associated protein |
| chr2_+_27498289 | 0.12 |
ENST00000296097.3
ENST00000420191.1 |
DNAJC5G
|
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma |
| chr1_-_183538319 | 0.12 |
ENST00000420553.1
ENST00000419402.1 |
NCF2
|
neutrophil cytosolic factor 2 |
| chr2_-_203735586 | 0.12 |
ENST00000454326.1
ENST00000432273.1 ENST00000450143.1 ENST00000411681.1 |
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr2_-_99871570 | 0.11 |
ENST00000333017.2
ENST00000409679.1 ENST00000423306.1 |
LYG2
|
lysozyme G-like 2 |
| chrM_+_10758 | 0.11 |
ENST00000361381.2
|
MT-ND4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr7_+_37723336 | 0.11 |
ENST00000450180.1
|
GPR141
|
G protein-coupled receptor 141 |
| chrX_+_134654540 | 0.11 |
ENST00000370752.4
|
DDX26B
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 26B |
| chrM_-_14670 | 0.11 |
ENST00000361681.2
|
MT-ND6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr1_-_92952433 | 0.11 |
ENST00000294702.5
|
GFI1
|
growth factor independent 1 transcription repressor |
| chr11_-_111649015 | 0.11 |
ENST00000529841.1
|
RP11-108O10.2
|
RP11-108O10.2 |
| chr2_-_198540751 | 0.11 |
ENST00000429081.1
|
RFTN2
|
raftlin family member 2 |
| chr1_+_199996702 | 0.11 |
ENST00000367362.3
|
NR5A2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr14_+_93389425 | 0.11 |
ENST00000216492.5
ENST00000334654.4 |
CHGA
|
chromogranin A (parathyroid secretory protein 1) |
| chr17_-_38928414 | 0.11 |
ENST00000335552.4
|
KRT26
|
keratin 26 |
| chrM_+_12331 | 0.11 |
ENST00000361567.2
|
MT-ND5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr2_-_145188137 | 0.10 |
ENST00000440875.1
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chrX_+_71401526 | 0.10 |
ENST00000218432.5
ENST00000423432.2 ENST00000373669.2 |
PIN4
|
protein (peptidylprolyl cis/trans isomerase) NIMA-interacting, 4 (parvulin) |
| chr19_-_7698599 | 0.10 |
ENST00000311069.5
|
PCP2
|
Purkinje cell protein 2 |
| chr7_-_99716914 | 0.10 |
ENST00000431404.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr12_+_78359999 | 0.10 |
ENST00000550503.1
|
NAV3
|
neuron navigator 3 |
| chr5_+_115358088 | 0.10 |
ENST00000600981.3
|
CTD-2287O16.3
|
CTD-2287O16.3 |
| chr8_-_112248400 | 0.10 |
ENST00000519506.1
ENST00000522778.1 |
RP11-946L20.4
|
RP11-946L20.4 |
| chr14_-_21737610 | 0.10 |
ENST00000320084.7
ENST00000449098.1 ENST00000336053.6 |
HNRNPC
|
heterogeneous nuclear ribonucleoprotein C (C1/C2) |
| chr5_+_125758865 | 0.10 |
ENST00000542322.1
ENST00000544396.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr14_-_73360796 | 0.10 |
ENST00000556509.1
ENST00000541685.1 ENST00000546183.1 |
DPF3
|
D4, zinc and double PHD fingers, family 3 |
| chr5_+_125758813 | 0.10 |
ENST00000285689.3
ENST00000515200.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr2_+_149402989 | 0.10 |
ENST00000397424.2
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr13_-_47471155 | 0.10 |
ENST00000543956.1
ENST00000542664.1 |
HTR2A
|
5-hydroxytryptamine (serotonin) receptor 2A, G protein-coupled |
| chr12_+_26348246 | 0.10 |
ENST00000422622.2
|
SSPN
|
sarcospan |
| chr4_+_169013666 | 0.10 |
ENST00000359299.3
|
ANXA10
|
annexin A10 |
| chr4_-_74486217 | 0.09 |
ENST00000335049.5
ENST00000307439.5 |
RASSF6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr14_-_23652849 | 0.09 |
ENST00000316902.7
ENST00000469263.1 ENST00000525062.1 ENST00000524758.1 |
SLC7A8
|
solute carrier family 7 (amino acid transporter light chain, L system), member 8 |
| chr10_-_23633720 | 0.09 |
ENST00000323327.4
|
C10orf67
|
chromosome 10 open reading frame 67 |
| chr7_-_99716952 | 0.09 |
ENST00000523306.1
ENST00000344095.4 ENST00000417349.1 ENST00000493322.1 ENST00000520135.1 ENST00000418432.2 ENST00000460673.2 ENST00000452041.1 ENST00000452438.2 ENST00000451699.1 ENST00000453269.2 |
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr18_-_47018869 | 0.09 |
ENST00000583036.1
ENST00000580261.1 |
RPL17
|
ribosomal protein L17 |
| chr18_-_47018769 | 0.09 |
ENST00000583637.1
ENST00000578528.1 ENST00000578532.1 ENST00000580387.1 ENST00000579248.1 ENST00000581373.1 |
RPL17
|
ribosomal protein L17 |
| chrX_-_13835147 | 0.09 |
ENST00000493677.1
ENST00000355135.2 |
GPM6B
|
glycoprotein M6B |
| chr17_+_73455788 | 0.09 |
ENST00000581519.1
|
KIAA0195
|
KIAA0195 |
| chr21_-_43816052 | 0.09 |
ENST00000398405.1
|
TMPRSS3
|
transmembrane protease, serine 3 |
| chr2_+_7118755 | 0.09 |
ENST00000433456.1
|
RNF144A
|
ring finger protein 144A |
| chr17_-_55911970 | 0.09 |
ENST00000581805.1
ENST00000580960.1 |
RP11-60A24.3
|
RP11-60A24.3 |
| chr14_+_52313833 | 0.09 |
ENST00000553560.1
|
GNG2
|
guanine nucleotide binding protein (G protein), gamma 2 |
| chr11_-_60010556 | 0.09 |
ENST00000427611.2
|
MS4A4E
|
membrane-spanning 4-domains, subfamily A, member 4E |
| chr14_+_21423611 | 0.09 |
ENST00000304625.2
|
RNASE2
|
ribonuclease, RNase A family, 2 (liver, eosinophil-derived neurotoxin) |
| chr4_+_95916947 | 0.09 |
ENST00000506363.1
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr2_+_12246664 | 0.09 |
ENST00000449986.1
|
AC096559.1
|
AC096559.1 |
| chr4_-_19458597 | 0.09 |
ENST00000505347.1
|
RP11-3J1.1
|
RP11-3J1.1 |
| chr18_+_3247779 | 0.09 |
ENST00000578611.1
ENST00000583449.1 |
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr4_-_170897045 | 0.09 |
ENST00000508313.1
|
RP11-205M3.3
|
RP11-205M3.3 |
| chr5_+_66300464 | 0.09 |
ENST00000436277.1
|
MAST4
|
microtubule associated serine/threonine kinase family member 4 |
| chr22_-_30960876 | 0.09 |
ENST00000401975.1
ENST00000428682.1 ENST00000423299.1 |
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr12_+_15699286 | 0.08 |
ENST00000442921.2
ENST00000542557.1 ENST00000445537.2 ENST00000544244.1 |
PTPRO
|
protein tyrosine phosphatase, receptor type, O |
| chr8_-_128231299 | 0.08 |
ENST00000500112.1
|
CCAT1
|
colon cancer associated transcript 1 (non-protein coding) |
| chr17_-_1532106 | 0.08 |
ENST00000301335.5
ENST00000382147.4 |
SLC43A2
|
solute carrier family 43 (amino acid system L transporter), member 2 |
| chr6_-_31107127 | 0.08 |
ENST00000259845.4
|
PSORS1C2
|
psoriasis susceptibility 1 candidate 2 |
| chr7_+_129015484 | 0.08 |
ENST00000490911.1
|
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr3_+_182983126 | 0.08 |
ENST00000481531.1
|
B3GNT5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chr2_-_145277569 | 0.08 |
ENST00000303660.4
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr1_-_149900122 | 0.08 |
ENST00000271628.8
|
SF3B4
|
splicing factor 3b, subunit 4, 49kDa |
| chr22_+_17956618 | 0.08 |
ENST00000262608.8
|
CECR2
|
cat eye syndrome chromosome region, candidate 2 |
| chr7_+_12726623 | 0.08 |
ENST00000439721.1
|
ARL4A
|
ADP-ribosylation factor-like 4A |
| chr15_+_41913690 | 0.08 |
ENST00000563576.1
|
MGA
|
MGA, MAX dimerization protein |
| chr2_-_102003987 | 0.08 |
ENST00000324768.5
|
CREG2
|
cellular repressor of E1A-stimulated genes 2 |
| chr3_-_193096600 | 0.08 |
ENST00000446087.1
ENST00000342358.4 |
ATP13A5
|
ATPase type 13A5 |
| chr6_-_109777128 | 0.08 |
ENST00000358807.3
ENST00000358577.3 |
MICAL1
|
microtubule associated monooxygenase, calponin and LIM domain containing 1 |
| chr10_+_18549645 | 0.08 |
ENST00000396576.2
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr16_-_29934558 | 0.08 |
ENST00000568995.1
ENST00000566413.1 |
KCTD13
|
potassium channel tetramerization domain containing 13 |
| chr12_+_128399965 | 0.08 |
ENST00000540882.1
ENST00000542089.1 |
LINC00507
|
long intergenic non-protein coding RNA 507 |
| chr19_-_7697857 | 0.08 |
ENST00000598935.1
|
PCP2
|
Purkinje cell protein 2 |
| chr1_+_160370344 | 0.08 |
ENST00000368061.2
|
VANGL2
|
VANGL planar cell polarity protein 2 |
| chr19_-_56110859 | 0.08 |
ENST00000221665.3
ENST00000592585.1 |
FIZ1
|
FLT3-interacting zinc finger 1 |
| chr15_+_58430567 | 0.08 |
ENST00000536493.1
|
AQP9
|
aquaporin 9 |
| chrX_-_77225135 | 0.08 |
ENST00000458128.1
|
PGAM4
|
phosphoglycerate mutase family member 4 |
| chr7_-_107883678 | 0.08 |
ENST00000417701.1
|
NRCAM
|
neuronal cell adhesion molecule |
| chrX_+_46696372 | 0.08 |
ENST00000218340.3
|
RP2
|
retinitis pigmentosa 2 (X-linked recessive) |
| chr7_-_99717463 | 0.08 |
ENST00000437822.2
|
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr6_+_31895287 | 0.08 |
ENST00000447952.2
|
C2
|
complement component 2 |
| chr8_-_29120604 | 0.08 |
ENST00000521515.1
|
KIF13B
|
kinesin family member 13B |
| chr17_-_28661065 | 0.08 |
ENST00000328886.4
ENST00000538566.2 |
TMIGD1
|
transmembrane and immunoglobulin domain containing 1 |
| chr6_-_127840336 | 0.08 |
ENST00000525778.1
|
SOGA3
|
SOGA family member 3 |
| chr1_-_190446759 | 0.08 |
ENST00000367462.3
|
BRINP3
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr5_-_78281775 | 0.08 |
ENST00000396151.3
ENST00000565165.1 |
ARSB
|
arylsulfatase B |
| chr18_-_47018897 | 0.07 |
ENST00000418495.1
|
RPL17
|
ribosomal protein L17 |
| chrX_-_110655306 | 0.07 |
ENST00000371993.2
|
DCX
|
doublecortin |
| chr20_+_42544782 | 0.07 |
ENST00000423191.2
ENST00000372999.1 |
TOX2
|
TOX high mobility group box family member 2 |
| chr13_+_78315348 | 0.07 |
ENST00000441784.1
|
SLAIN1
|
SLAIN motif family, member 1 |
| chr3_+_173116225 | 0.07 |
ENST00000457714.1
|
NLGN1
|
neuroligin 1 |
| chr2_-_69664586 | 0.07 |
ENST00000303698.3
ENST00000394305.1 ENST00000410022.2 |
NFU1
|
NFU1 iron-sulfur cluster scaffold homolog (S. cerevisiae) |
| chr15_+_58430368 | 0.07 |
ENST00000558772.1
ENST00000219919.4 |
AQP9
|
aquaporin 9 |
| chr19_+_50180409 | 0.07 |
ENST00000391851.4
|
PRMT1
|
protein arginine methyltransferase 1 |
| chr13_-_36050819 | 0.07 |
ENST00000379919.4
|
MAB21L1
|
mab-21-like 1 (C. elegans) |
| chrX_-_15332665 | 0.07 |
ENST00000537676.1
ENST00000344384.4 |
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chr2_+_234826016 | 0.07 |
ENST00000324695.4
ENST00000433712.2 |
TRPM8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr17_-_46690839 | 0.07 |
ENST00000498634.2
|
HOXB8
|
homeobox B8 |
| chr1_+_38478378 | 0.07 |
ENST00000373014.4
|
UTP11L
|
UTP11-like, U3 small nucleolar ribonucleoprotein, (yeast) |
| chr10_-_21186144 | 0.07 |
ENST00000377119.1
|
NEBL
|
nebulette |
| chrX_-_14047996 | 0.07 |
ENST00000380523.4
ENST00000398355.3 |
GEMIN8
|
gem (nuclear organelle) associated protein 8 |
| chrX_-_21676442 | 0.07 |
ENST00000379499.2
|
KLHL34
|
kelch-like family member 34 |
| chr8_+_77593448 | 0.07 |
ENST00000521891.2
|
ZFHX4
|
zinc finger homeobox 4 |
| chr20_+_45947246 | 0.07 |
ENST00000599904.1
|
AL031666.2
|
HCG2018772; Uncharacterized protein; cDNA FLJ31609 fis, clone NT2RI2002852 |
| chr11_+_65554493 | 0.07 |
ENST00000335987.3
|
OVOL1
|
ovo-like zinc finger 1 |
| chr4_+_8201091 | 0.07 |
ENST00000382521.3
ENST00000245105.3 ENST00000457650.2 ENST00000539824.1 |
SH3TC1
|
SH3 domain and tetratricopeptide repeats 1 |
| chr9_-_107690420 | 0.07 |
ENST00000423487.2
ENST00000374733.1 ENST00000374736.3 |
ABCA1
|
ATP-binding cassette, sub-family A (ABC1), member 1 |
| chr17_-_7145475 | 0.07 |
ENST00000571129.1
ENST00000571253.1 ENST00000573928.1 |
GABARAP
|
GABA(A) receptor-associated protein |
| chr17_+_12569306 | 0.07 |
ENST00000425538.1
|
MYOCD
|
myocardin |
| chr7_-_100860851 | 0.07 |
ENST00000223127.3
|
PLOD3
|
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
| chr21_+_33671160 | 0.07 |
ENST00000303645.5
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr12_-_6233828 | 0.07 |
ENST00000572068.1
ENST00000261405.5 |
VWF
|
von Willebrand factor |
| chr6_+_127898312 | 0.07 |
ENST00000329722.7
|
C6orf58
|
chromosome 6 open reading frame 58 |
| chr16_+_1359511 | 0.07 |
ENST00000397514.3
ENST00000397515.2 ENST00000567383.1 ENST00000403747.2 ENST00000566587.1 |
UBE2I
|
ubiquitin-conjugating enzyme E2I |
| chr3_-_99595037 | 0.07 |
ENST00000383694.2
|
FILIP1L
|
filamin A interacting protein 1-like |
| chr14_+_52164820 | 0.07 |
ENST00000554167.1
|
FRMD6
|
FERM domain containing 6 |
| chr10_-_50970322 | 0.07 |
ENST00000374103.4
|
OGDHL
|
oxoglutarate dehydrogenase-like |
| chr12_-_94673956 | 0.07 |
ENST00000551941.1
|
RP11-1105G2.3
|
Uncharacterized protein |
| chr19_+_15838834 | 0.06 |
ENST00000305899.3
|
OR10H2
|
olfactory receptor, family 10, subfamily H, member 2 |
| chrX_+_36254051 | 0.06 |
ENST00000378657.4
|
CXorf30
|
chromosome X open reading frame 30 |
| chr19_-_58951496 | 0.06 |
ENST00000254166.3
|
ZNF132
|
zinc finger protein 132 |
| chr13_-_30880979 | 0.06 |
ENST00000414289.1
|
KATNAL1
|
katanin p60 subunit A-like 1 |
| chr7_+_128502895 | 0.06 |
ENST00000492758.1
|
ATP6V1F
|
ATPase, H+ transporting, lysosomal 14kDa, V1 subunit F |
| chr18_+_22040620 | 0.06 |
ENST00000426880.2
|
HRH4
|
histamine receptor H4 |
| chr12_+_128399917 | 0.06 |
ENST00000544645.1
|
LINC00507
|
long intergenic non-protein coding RNA 507 |
| chr3_-_99594948 | 0.06 |
ENST00000471562.1
ENST00000495625.2 |
FILIP1L
|
filamin A interacting protein 1-like |
| chr12_-_10605929 | 0.06 |
ENST00000347831.5
ENST00000359151.3 |
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1 |
| chr17_+_79650962 | 0.06 |
ENST00000329138.4
|
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate |
| chr5_+_174151536 | 0.06 |
ENST00000239243.6
ENST00000507785.1 |
MSX2
|
msh homeobox 2 |
| chr11_-_26593649 | 0.06 |
ENST00000455601.2
|
MUC15
|
mucin 15, cell surface associated |
| chr2_+_210636697 | 0.06 |
ENST00000439458.1
ENST00000272845.6 |
UNC80
|
unc-80 homolog (C. elegans) |
| chr17_+_33914424 | 0.06 |
ENST00000590432.1
|
AP2B1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr3_-_157217328 | 0.06 |
ENST00000392832.2
ENST00000543418.1 |
VEPH1
|
ventricular zone expressed PH domain-containing 1 |
| chr2_+_202047843 | 0.06 |
ENST00000272879.5
ENST00000374650.3 ENST00000346817.5 ENST00000313728.7 ENST00000448480.1 |
CASP10
|
caspase 10, apoptosis-related cysteine peptidase |
| chr4_-_89205879 | 0.06 |
ENST00000608933.1
ENST00000315194.4 ENST00000514204.1 |
PPM1K
|
protein phosphatase, Mg2+/Mn2+ dependent, 1K |
| chr2_-_214016314 | 0.06 |
ENST00000434687.1
ENST00000374319.4 |
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chrX_+_105855160 | 0.06 |
ENST00000372544.2
ENST00000372548.4 |
CXorf57
|
chromosome X open reading frame 57 |
| chr7_+_128502871 | 0.06 |
ENST00000249289.4
|
ATP6V1F
|
ATPase, H+ transporting, lysosomal 14kDa, V1 subunit F |
| chr14_+_92789498 | 0.06 |
ENST00000531433.1
|
SLC24A4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
| chr20_-_45976816 | 0.06 |
ENST00000441977.1
|
ZMYND8
|
zinc finger, MYND-type containing 8 |
| chr11_-_107729887 | 0.06 |
ENST00000525815.1
|
SLC35F2
|
solute carrier family 35, member F2 |
| chr17_+_46970127 | 0.06 |
ENST00000355938.5
|
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr12_-_16760195 | 0.06 |
ENST00000546281.1
ENST00000537757.1 |
LMO3
|
LIM domain only 3 (rhombotin-like 2) |
| chr1_+_160160283 | 0.06 |
ENST00000368079.3
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr8_-_49834299 | 0.06 |
ENST00000396822.1
|
SNAI2
|
snail family zinc finger 2 |
| chr12_-_95945246 | 0.06 |
ENST00000258499.3
|
USP44
|
ubiquitin specific peptidase 44 |
| chr11_+_119076745 | 0.06 |
ENST00000264033.4
|
CBL
|
Cbl proto-oncogene, E3 ubiquitin protein ligase |
| chr12_+_64798826 | 0.06 |
ENST00000540203.1
|
XPOT
|
exportin, tRNA |
| chr5_+_140602904 | 0.06 |
ENST00000515856.2
ENST00000239449.4 |
PCDHB14
|
protocadherin beta 14 |
Network of associatons between targets according to the STRING database.
First level regulatory network of EMX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.1 | 0.2 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.1 | 0.2 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.0 | 0.2 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.0 | 0.3 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.7 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.2 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.2 | GO:1904823 | pyrimidine nucleobase transport(GO:0015855) purine nucleobase transmembrane transport(GO:1904823) |
| 0.0 | 0.1 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.1 | GO:1900738 | dense core granule biogenesis(GO:0061110) positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) positive regulation of relaxation of cardiac muscle(GO:1901899) regulation of dense core granule biogenesis(GO:2000705) |
| 0.0 | 0.2 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.1 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.1 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.1 | GO:0000294 | nuclear-transcribed mRNA catabolic process, endonucleolytic cleavage-dependent decay(GO:0000294) |
| 0.0 | 0.1 | GO:0060489 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.0 | 0.1 | GO:0061580 | colon epithelial cell migration(GO:0061580) |
| 0.0 | 0.1 | GO:0007070 | negative regulation of transcription during mitosis(GO:0007068) negative regulation of transcription from RNA polymerase II promoter during mitosis(GO:0007070) |
| 0.0 | 0.1 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.0 | 0.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.4 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.1 | GO:1901994 | negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.2 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.1 | GO:0051795 | positive regulation of catagen(GO:0051795) activation of meiosis(GO:0090427) |
| 0.0 | 0.2 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.1 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.1 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.1 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.0 | 0.1 | GO:1902231 | positive regulation of keratinocyte apoptotic process(GO:1902174) positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.1 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.1 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.2 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.0 | GO:0051801 | cytolysis by symbiont of host cells(GO:0001897) cytolysis in other organism involved in symbiotic interaction(GO:0051801) |
| 0.0 | 0.0 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.0 | 0.0 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.0 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.0 | 0.2 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.0 | GO:1904204 | skeletal muscle hypertrophy(GO:0014734) regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.0 | 0.0 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.0 | GO:0032641 | lymphotoxin A production(GO:0032641) lymphotoxin A biosynthetic process(GO:0042109) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.0 | 0.1 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 0.0 | 0.1 | GO:0032489 | aminophospholipid transport(GO:0015917) regulation of Cdc42 protein signal transduction(GO:0032489) regulation of high-density lipoprotein particle assembly(GO:0090107) |
| 0.0 | 0.1 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.2 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.1 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.0 | 0.1 | GO:0030807 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.0 | 0.2 | GO:0014877 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.0 | GO:0060382 | regulation of DNA strand elongation(GO:0060382) |
| 0.0 | 0.4 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.1 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.0 | 0.1 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.3 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.0 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.0 | 0.2 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.0 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.0 | 0.3 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.1 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.3 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.2 | GO:0097179 | protease inhibitor complex(GO:0097179) |
| 0.0 | 0.4 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.1 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.0 | 0.1 | GO:0043614 | multi-eIF complex(GO:0043614) translation preinitiation complex(GO:0070993) astrocyte end-foot(GO:0097450) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.1 | GO:0033565 | ESCRT-0 complex(GO:0033565) |
| 0.0 | 0.1 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.0 | 0.1 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.0 | 0.0 | GO:0044218 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 0.2 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.3 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.2 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.2 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.1 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.1 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.1 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 0.2 | GO:0015265 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) glycerol channel activity(GO:0015254) urea channel activity(GO:0015265) |
| 0.0 | 0.1 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.1 | GO:0031783 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.1 | GO:0008457 | beta-galactosyl-N-acetylglucosaminylgalactosylglucosyl-ceramide beta-1,3-acetylglucosaminyltransferase activity(GO:0008457) lactosylceramide 1,3-N-acetyl-beta-D-glucosaminyltransferase activity(GO:0047256) |
| 0.0 | 0.2 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.4 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.4 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.3 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.2 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.1 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.0 | 0.1 | GO:0034188 | apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.0 | 0.1 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.1 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.0 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.0 | GO:0015633 | zinc transporting ATPase activity(GO:0015633) |
| 0.0 | 0.0 | GO:0005333 | acetylcholine transmembrane transporter activity(GO:0005277) norepinephrine transmembrane transporter activity(GO:0005333) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.0 | 0.0 | GO:1904854 | proteasome core complex binding(GO:1904854) |
| 0.0 | 0.1 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.2 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.1 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.1 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.1 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.0 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.0 | 0.0 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 0.0 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.3 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.2 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |