Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
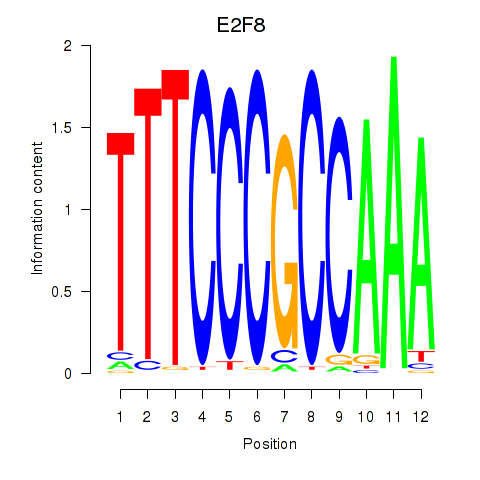
Results for E2F8
Z-value: 0.65
Transcription factors associated with E2F8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
E2F8
|
ENSG00000129173.8 | E2F transcription factor 8 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E2F8 | hg19_v2_chr11_-_19263145_19263176 | -0.87 | 1.3e-01 | Click! |
Activity profile of E2F8 motif
Sorted Z-values of E2F8 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_+_150866779 | 1.25 |
ENST00000370353.3
|
PRRG3
|
proline rich Gla (G-carboxyglutamic acid) 3 (transmembrane) |
| chr2_+_232575168 | 0.45 |
ENST00000440384.1
|
PTMA
|
prothymosin, alpha |
| chr4_-_176733897 | 0.39 |
ENST00000393658.2
|
GPM6A
|
glycoprotein M6A |
| chr19_+_51153045 | 0.34 |
ENST00000458538.1
|
C19orf81
|
chromosome 19 open reading frame 81 |
| chr1_+_153330322 | 0.34 |
ENST00000368738.3
|
S100A9
|
S100 calcium binding protein A9 |
| chr6_-_85473073 | 0.33 |
ENST00000606621.1
|
TBX18
|
T-box 18 |
| chr1_+_158979792 | 0.33 |
ENST00000359709.3
ENST00000430894.2 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr14_-_67878917 | 0.29 |
ENST00000216446.4
|
PLEK2
|
pleckstrin 2 |
| chr4_+_25314388 | 0.28 |
ENST00000302874.4
|
ZCCHC4
|
zinc finger, CCHC domain containing 4 |
| chr6_-_84937314 | 0.26 |
ENST00000257766.4
ENST00000403245.3 |
KIAA1009
|
KIAA1009 |
| chr4_-_140223614 | 0.25 |
ENST00000394223.1
|
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr19_-_51845378 | 0.24 |
ENST00000335624.4
|
VSIG10L
|
V-set and immunoglobulin domain containing 10 like |
| chr4_-_140223670 | 0.23 |
ENST00000394228.1
ENST00000539387.1 |
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr11_-_327537 | 0.22 |
ENST00000602735.1
|
IFITM3
|
interferon induced transmembrane protein 3 |
| chr14_-_51135005 | 0.21 |
ENST00000556735.1
|
SAV1
|
salvador homolog 1 (Drosophila) |
| chr20_+_31595406 | 0.21 |
ENST00000170150.3
|
BPIFB2
|
BPI fold containing family B, member 2 |
| chr14_+_45553296 | 0.21 |
ENST00000355765.6
ENST00000553605.1 |
PRPF39
|
pre-mRNA processing factor 39 |
| chr4_-_129209221 | 0.21 |
ENST00000512483.1
|
PGRMC2
|
progesterone receptor membrane component 2 |
| chr17_-_62499334 | 0.21 |
ENST00000579996.1
|
DDX5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr7_+_114562616 | 0.20 |
ENST00000448022.1
|
MDFIC
|
MyoD family inhibitor domain containing |
| chr2_-_152146385 | 0.20 |
ENST00000414946.1
ENST00000243346.5 |
NMI
|
N-myc (and STAT) interactor |
| chr11_-_71639613 | 0.20 |
ENST00000528184.1
ENST00000528511.2 |
RP11-849H4.2
|
Putative short transient receptor potential channel 2-like protein |
| chr18_-_47018869 | 0.20 |
ENST00000583036.1
ENST00000580261.1 |
RPL17
|
ribosomal protein L17 |
| chr1_+_197871854 | 0.20 |
ENST00000436652.1
|
C1orf53
|
chromosome 1 open reading frame 53 |
| chr3_+_122296465 | 0.20 |
ENST00000483793.1
|
PARP15
|
poly (ADP-ribose) polymerase family, member 15 |
| chr9_+_8858102 | 0.20 |
ENST00000447950.1
ENST00000430766.1 |
RP11-75C9.1
|
RP11-75C9.1 |
| chr6_+_135502408 | 0.19 |
ENST00000341911.5
ENST00000442647.2 ENST00000316528.8 |
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr19_+_41103063 | 0.19 |
ENST00000308370.7
|
LTBP4
|
latent transforming growth factor beta binding protein 4 |
| chr6_-_26659913 | 0.18 |
ENST00000480036.1
ENST00000415922.2 |
ZNF322
|
zinc finger protein 322 |
| chr11_+_17298255 | 0.18 |
ENST00000531172.1
ENST00000533738.2 ENST00000323688.6 |
NUCB2
|
nucleobindin 2 |
| chr17_+_6915730 | 0.18 |
ENST00000548577.1
|
RNASEK
|
ribonuclease, RNase K |
| chr2_-_24346218 | 0.18 |
ENST00000436622.1
ENST00000313213.4 |
PFN4
|
profilin family, member 4 |
| chr3_-_54962100 | 0.18 |
ENST00000273286.5
|
LRTM1
|
leucine-rich repeats and transmembrane domains 1 |
| chr7_+_114562909 | 0.18 |
ENST00000423503.1
ENST00000427207.1 |
MDFIC
|
MyoD family inhibitor domain containing |
| chr14_-_68157084 | 0.18 |
ENST00000557564.1
|
RP11-1012A1.4
|
RP11-1012A1.4 |
| chr11_+_17298297 | 0.17 |
ENST00000529010.1
|
NUCB2
|
nucleobindin 2 |
| chr17_+_6915798 | 0.17 |
ENST00000402093.1
|
RNASEK
|
ribonuclease, RNase K |
| chr17_+_54671047 | 0.17 |
ENST00000332822.4
|
NOG
|
noggin |
| chr8_+_29952914 | 0.17 |
ENST00000321250.8
ENST00000518001.1 ENST00000520682.1 ENST00000442880.2 ENST00000523116.1 |
LEPROTL1
|
leptin receptor overlapping transcript-like 1 |
| chr2_+_231191875 | 0.17 |
ENST00000444636.1
ENST00000415673.2 ENST00000243810.6 ENST00000396563.4 |
SP140L
|
SP140 nuclear body protein-like |
| chr6_+_160693591 | 0.17 |
ENST00000419196.1
|
RP1-276N6.2
|
RP1-276N6.2 |
| chr12_+_57146233 | 0.17 |
ENST00000554643.1
ENST00000556650.1 ENST00000554150.1 ENST00000554155.1 |
HSD17B6
|
hydroxysteroid (17-beta) dehydrogenase 6 |
| chr3_+_70048881 | 0.16 |
ENST00000483525.1
|
RP11-460N16.1
|
RP11-460N16.1 |
| chr9_-_117267717 | 0.16 |
ENST00000374057.3
|
DFNB31
|
deafness, autosomal recessive 31 |
| chr2_+_109745655 | 0.16 |
ENST00000418513.1
|
SH3RF3
|
SH3 domain containing ring finger 3 |
| chr16_+_56691911 | 0.16 |
ENST00000568475.1
|
MT1F
|
metallothionein 1F |
| chr6_-_57086200 | 0.16 |
ENST00000468148.1
|
RAB23
|
RAB23, member RAS oncogene family |
| chr21_+_37477164 | 0.16 |
ENST00000422473.1
|
AP000688.29
|
AP000688.29 |
| chr8_+_145065705 | 0.16 |
ENST00000533044.1
|
GRINA
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chr9_+_129097520 | 0.16 |
ENST00000436593.3
|
MVB12B
|
multivesicular body subunit 12B |
| chr14_+_102027688 | 0.16 |
ENST00000510508.4
ENST00000359323.3 |
DIO3
|
deiodinase, iodothyronine, type III |
| chr17_-_4463856 | 0.16 |
ENST00000574584.1
ENST00000381550.3 ENST00000301395.3 |
GGT6
|
gamma-glutamyltransferase 6 |
| chr15_+_44719996 | 0.15 |
ENST00000559793.1
ENST00000558968.1 |
CTDSPL2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr14_+_77582905 | 0.15 |
ENST00000557408.1
|
TMEM63C
|
transmembrane protein 63C |
| chr12_-_123011476 | 0.15 |
ENST00000528279.1
ENST00000344591.4 ENST00000526560.2 |
RSRC2
|
arginine/serine-rich coiled-coil 2 |
| chr17_-_56084578 | 0.15 |
ENST00000582730.2
ENST00000584773.1 ENST00000585096.1 ENST00000258962.4 |
SRSF1
|
serine/arginine-rich splicing factor 1 |
| chr1_+_92632542 | 0.15 |
ENST00000409154.4
ENST00000370378.4 |
KIAA1107
|
KIAA1107 |
| chr8_+_69242957 | 0.15 |
ENST00000518698.1
ENST00000539993.1 |
C8orf34
|
chromosome 8 open reading frame 34 |
| chr3_+_14058794 | 0.14 |
ENST00000424053.1
ENST00000528067.1 ENST00000429201.1 |
TPRXL
|
tetra-peptide repeat homeobox-like |
| chr3_+_14474178 | 0.14 |
ENST00000452775.1
|
SLC6A6
|
solute carrier family 6 (neurotransmitter transporter), member 6 |
| chr5_+_31532373 | 0.14 |
ENST00000325366.9
ENST00000355907.3 ENST00000507818.2 |
C5orf22
|
chromosome 5 open reading frame 22 |
| chr1_+_33283043 | 0.14 |
ENST00000373476.1
ENST00000373475.5 ENST00000529027.1 ENST00000398243.3 |
S100PBP
|
S100P binding protein |
| chr5_+_133707252 | 0.14 |
ENST00000506787.1
ENST00000507277.1 |
UBE2B
|
ubiquitin-conjugating enzyme E2B |
| chr12_-_71003568 | 0.14 |
ENST00000547715.1
ENST00000451516.2 ENST00000538708.1 ENST00000550857.1 ENST00000261266.5 |
PTPRB
|
protein tyrosine phosphatase, receptor type, B |
| chr1_+_244998918 | 0.14 |
ENST00000366528.3
|
COX20
|
COX20 cytochrome C oxidase assembly factor |
| chr12_+_111843749 | 0.14 |
ENST00000341259.2
|
SH2B3
|
SH2B adaptor protein 3 |
| chr3_-_187455680 | 0.14 |
ENST00000438077.1
|
BCL6
|
B-cell CLL/lymphoma 6 |
| chr3_-_25824925 | 0.13 |
ENST00000396649.3
ENST00000428257.1 ENST00000280700.5 |
NGLY1
|
N-glycanase 1 |
| chr17_-_60005329 | 0.13 |
ENST00000251334.6
|
INTS2
|
integrator complex subunit 2 |
| chr19_+_49977466 | 0.13 |
ENST00000596435.1
ENST00000344019.3 ENST00000597551.1 ENST00000204637.2 ENST00000600429.1 |
FLT3LG
|
fms-related tyrosine kinase 3 ligand |
| chr15_-_34635314 | 0.13 |
ENST00000557912.1
ENST00000328848.4 |
NOP10
|
NOP10 ribonucleoprotein |
| chr2_+_130939235 | 0.13 |
ENST00000425361.1
ENST00000457492.1 |
MZT2B
|
mitotic spindle organizing protein 2B |
| chr19_+_57874835 | 0.13 |
ENST00000543226.1
ENST00000596755.1 ENST00000282282.3 ENST00000597658.1 |
TRAPPC2P1
ZNF547
AC003002.4
|
trafficking protein particle complex 2 pseudogene 1 zinc finger protein 547 Uncharacterized protein |
| chr14_-_69619291 | 0.13 |
ENST00000554215.1
ENST00000556847.1 |
DCAF5
|
DDB1 and CUL4 associated factor 5 |
| chr4_-_130014729 | 0.13 |
ENST00000281142.5
ENST00000434680.1 |
SCLT1
|
sodium channel and clathrin linker 1 |
| chr17_-_7155775 | 0.13 |
ENST00000571409.1
|
CTDNEP1
|
CTD nuclear envelope phosphatase 1 |
| chr6_+_24357131 | 0.12 |
ENST00000274766.1
|
KAAG1
|
kidney associated antigen 1 |
| chr2_+_16080659 | 0.12 |
ENST00000281043.3
|
MYCN
|
v-myc avian myelocytomatosis viral oncogene neuroblastoma derived homolog |
| chr19_-_49828438 | 0.12 |
ENST00000454748.3
ENST00000598828.1 ENST00000335875.4 |
SLC6A16
|
solute carrier family 6, member 16 |
| chr8_-_112248400 | 0.12 |
ENST00000519506.1
ENST00000522778.1 |
RP11-946L20.4
|
RP11-946L20.4 |
| chr17_+_6915902 | 0.12 |
ENST00000570898.1
ENST00000552842.1 |
RNASEK
|
ribonuclease, RNase K |
| chr18_-_47018897 | 0.12 |
ENST00000418495.1
|
RPL17
|
ribosomal protein L17 |
| chr1_+_70876926 | 0.12 |
ENST00000370938.3
ENST00000346806.2 |
CTH
|
cystathionase (cystathionine gamma-lyase) |
| chr8_+_118533049 | 0.12 |
ENST00000522839.1
|
MED30
|
mediator complex subunit 30 |
| chr3_+_171758344 | 0.12 |
ENST00000336824.4
ENST00000423424.1 |
FNDC3B
|
fibronectin type III domain containing 3B |
| chr12_-_110939870 | 0.12 |
ENST00000447578.2
ENST00000546588.1 ENST00000360579.7 ENST00000549970.1 ENST00000549578.1 |
VPS29
|
vacuolar protein sorting 29 homolog (S. cerevisiae) |
| chr1_-_150693305 | 0.12 |
ENST00000368987.1
|
HORMAD1
|
HORMA domain containing 1 |
| chr2_-_113993020 | 0.12 |
ENST00000465084.1
|
PAX8
|
paired box 8 |
| chr22_+_20104947 | 0.12 |
ENST00000402752.1
|
RANBP1
|
RAN binding protein 1 |
| chr8_-_131455835 | 0.11 |
ENST00000518721.1
|
ASAP1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
| chr4_+_128702969 | 0.11 |
ENST00000508776.1
ENST00000439123.2 |
HSPA4L
|
heat shock 70kDa protein 4-like |
| chr1_+_32827759 | 0.11 |
ENST00000373534.3
|
TSSK3
|
testis-specific serine kinase 3 |
| chr2_-_37193606 | 0.11 |
ENST00000379213.2
ENST00000263918.4 |
STRN
|
striatin, calmodulin binding protein |
| chr15_-_100258029 | 0.11 |
ENST00000378904.2
|
DKFZP779J2370
|
DKFZP779J2370 |
| chrX_-_122866874 | 0.11 |
ENST00000245838.8
ENST00000355725.4 |
THOC2
|
THO complex 2 |
| chr12_+_56324756 | 0.11 |
ENST00000331886.5
ENST00000555090.1 |
DGKA
|
diacylglycerol kinase, alpha 80kDa |
| chr5_+_34757309 | 0.11 |
ENST00000397449.1
|
RAI14
|
retinoic acid induced 14 |
| chr2_+_148778570 | 0.11 |
ENST00000407073.1
|
MBD5
|
methyl-CpG binding domain protein 5 |
| chr9_+_79792410 | 0.11 |
ENST00000357409.5
|
VPS13A
|
vacuolar protein sorting 13 homolog A (S. cerevisiae) |
| chr10_+_106028923 | 0.11 |
ENST00000338595.2
|
GSTO2
|
glutathione S-transferase omega 2 |
| chr9_-_96717654 | 0.11 |
ENST00000253968.6
|
BARX1
|
BARX homeobox 1 |
| chr11_-_71639446 | 0.11 |
ENST00000534704.1
|
RP11-849H4.2
|
Putative short transient receptor potential channel 2-like protein |
| chr6_+_30034966 | 0.11 |
ENST00000376769.2
|
PPP1R11
|
protein phosphatase 1, regulatory (inhibitor) subunit 11 |
| chr18_+_29078131 | 0.11 |
ENST00000585206.1
|
DSG2
|
desmoglein 2 |
| chr8_+_128426535 | 0.11 |
ENST00000465342.2
|
POU5F1B
|
POU class 5 homeobox 1B |
| chr6_+_154360616 | 0.11 |
ENST00000229768.5
ENST00000419506.2 ENST00000524163.1 ENST00000414028.2 ENST00000435918.2 ENST00000337049.4 |
OPRM1
|
opioid receptor, mu 1 |
| chr11_+_35201826 | 0.10 |
ENST00000531873.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr1_+_205473720 | 0.10 |
ENST00000429964.2
ENST00000506784.1 ENST00000360066.2 |
CDK18
|
cyclin-dependent kinase 18 |
| chr7_+_116593292 | 0.10 |
ENST00000393446.2
ENST00000265437.5 ENST00000393451.3 |
ST7
|
suppression of tumorigenicity 7 |
| chr9_+_129097479 | 0.10 |
ENST00000402437.2
|
MVB12B
|
multivesicular body subunit 12B |
| chr15_-_93965805 | 0.10 |
ENST00000556708.1
|
RP11-164C12.2
|
RP11-164C12.2 |
| chr8_+_27632083 | 0.10 |
ENST00000519637.1
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr3_-_44803141 | 0.10 |
ENST00000296121.4
|
KIAA1143
|
KIAA1143 |
| chr15_-_60884706 | 0.10 |
ENST00000449337.2
|
RORA
|
RAR-related orphan receptor A |
| chr14_-_51135036 | 0.10 |
ENST00000324679.4
|
SAV1
|
salvador homolog 1 (Drosophila) |
| chr12_+_56324933 | 0.10 |
ENST00000549629.1
ENST00000555218.1 |
DGKA
|
diacylglycerol kinase, alpha 80kDa |
| chr12_-_123011536 | 0.10 |
ENST00000331738.7
ENST00000354654.2 |
RSRC2
|
arginine/serine-rich coiled-coil 2 |
| chr15_+_44719394 | 0.09 |
ENST00000260327.4
ENST00000396780.1 |
CTDSPL2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr11_+_126225529 | 0.09 |
ENST00000227495.6
ENST00000444328.2 ENST00000356132.4 |
ST3GAL4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr11_-_73882249 | 0.09 |
ENST00000535954.1
|
C2CD3
|
C2 calcium-dependent domain containing 3 |
| chr17_+_5389605 | 0.09 |
ENST00000576988.1
ENST00000576570.1 ENST00000573759.1 |
MIS12
|
MIS12 kinetochore complex component |
| chr14_-_69619689 | 0.09 |
ENST00000389997.6
ENST00000557386.1 ENST00000554681.1 |
DCAF5
|
DDB1 and CUL4 associated factor 5 |
| chr18_-_47813940 | 0.09 |
ENST00000586837.1
ENST00000412036.2 ENST00000589940.1 |
CXXC1
|
CXXC finger protein 1 |
| chr14_-_69619823 | 0.09 |
ENST00000341516.5
|
DCAF5
|
DDB1 and CUL4 associated factor 5 |
| chr7_+_100136811 | 0.09 |
ENST00000300176.4
ENST00000262935.4 |
AGFG2
|
ArfGAP with FG repeats 2 |
| chr11_+_118868830 | 0.09 |
ENST00000334418.1
|
CCDC84
|
coiled-coil domain containing 84 |
| chr13_+_32889605 | 0.09 |
ENST00000380152.3
ENST00000544455.1 ENST00000530893.2 |
BRCA2
|
breast cancer 2, early onset |
| chr8_+_145065521 | 0.09 |
ENST00000534791.1
|
GRINA
|
glutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) |
| chr1_-_21503337 | 0.08 |
ENST00000400422.1
ENST00000602326.1 ENST00000411888.1 ENST00000438975.1 |
EIF4G3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr6_+_144164455 | 0.08 |
ENST00000367576.5
|
LTV1
|
LTV1 homolog (S. cerevisiae) |
| chr1_-_28241226 | 0.08 |
ENST00000373912.3
ENST00000373909.3 |
RPA2
|
replication protein A2, 32kDa |
| chr21_+_19273574 | 0.08 |
ENST00000400128.1
|
CHODL
|
chondrolectin |
| chr2_+_230787201 | 0.08 |
ENST00000283946.3
|
FBXO36
|
F-box protein 36 |
| chr11_+_125496124 | 0.08 |
ENST00000533778.2
ENST00000534070.1 |
CHEK1
|
checkpoint kinase 1 |
| chrX_-_153210107 | 0.08 |
ENST00000369997.3
ENST00000393700.3 ENST00000412763.1 |
RENBP
|
renin binding protein |
| chr15_+_44719790 | 0.08 |
ENST00000558791.1
|
CTDSPL2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase like 2 |
| chr2_-_198650037 | 0.08 |
ENST00000392296.4
|
BOLL
|
boule-like RNA-binding protein |
| chr8_-_92053042 | 0.08 |
ENST00000520014.1
|
TMEM55A
|
transmembrane protein 55A |
| chr4_-_129209944 | 0.08 |
ENST00000520121.1
|
PGRMC2
|
progesterone receptor membrane component 2 |
| chr16_-_31147020 | 0.08 |
ENST00000568261.1
ENST00000567797.1 ENST00000317508.6 |
PRSS8
|
protease, serine, 8 |
| chr17_+_57642886 | 0.08 |
ENST00000251241.4
ENST00000451169.2 ENST00000425628.3 ENST00000584385.1 ENST00000580030.1 |
DHX40
|
DEAH (Asp-Glu-Ala-His) box polypeptide 40 |
| chr9_+_130374537 | 0.08 |
ENST00000373302.3
ENST00000373299.1 |
STXBP1
|
syntaxin binding protein 1 |
| chrX_-_119695279 | 0.08 |
ENST00000336592.6
|
CUL4B
|
cullin 4B |
| chr15_-_57025675 | 0.08 |
ENST00000558320.1
|
ZNF280D
|
zinc finger protein 280D |
| chr14_-_20801427 | 0.08 |
ENST00000557665.1
ENST00000358932.4 ENST00000353689.4 |
CCNB1IP1
|
cyclin B1 interacting protein 1, E3 ubiquitin protein ligase |
| chr11_-_71639670 | 0.08 |
ENST00000533047.1
ENST00000529844.1 |
RP11-849H4.2
|
Putative short transient receptor potential channel 2-like protein |
| chr19_+_57791806 | 0.07 |
ENST00000360338.3
|
ZNF460
|
zinc finger protein 460 |
| chr3_-_79816965 | 0.07 |
ENST00000464233.1
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr6_+_36165133 | 0.07 |
ENST00000446974.1
ENST00000454960.1 |
BRPF3
|
bromodomain and PHD finger containing, 3 |
| chr15_-_40074996 | 0.07 |
ENST00000350221.3
|
FSIP1
|
fibrous sheath interacting protein 1 |
| chr11_+_47586982 | 0.07 |
ENST00000426530.2
ENST00000534775.1 |
PTPMT1
|
protein tyrosine phosphatase, mitochondrial 1 |
| chr10_-_95360983 | 0.07 |
ENST00000371464.3
|
RBP4
|
retinol binding protein 4, plasma |
| chr12_-_82752159 | 0.07 |
ENST00000552377.1
|
CCDC59
|
coiled-coil domain containing 59 |
| chr7_-_152373216 | 0.07 |
ENST00000359321.1
|
XRCC2
|
X-ray repair complementing defective repair in Chinese hamster cells 2 |
| chr3_-_57113281 | 0.07 |
ENST00000468466.1
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3 |
| chr2_-_148778323 | 0.07 |
ENST00000440042.1
ENST00000535373.1 ENST00000540442.1 ENST00000536575.1 |
ORC4
|
origin recognition complex, subunit 4 |
| chrX_-_130037162 | 0.07 |
ENST00000432489.1
|
ENOX2
|
ecto-NOX disulfide-thiol exchanger 2 |
| chr8_+_29953163 | 0.07 |
ENST00000518192.1
|
LEPROTL1
|
leptin receptor overlapping transcript-like 1 |
| chr14_+_55595762 | 0.07 |
ENST00000254301.9
|
LGALS3
|
lectin, galactoside-binding, soluble, 3 |
| chrX_+_133371077 | 0.07 |
ENST00000517294.1
ENST00000370809.4 |
CCDC160
|
coiled-coil domain containing 160 |
| chr5_+_142125161 | 0.07 |
ENST00000432677.1
|
AC005592.1
|
AC005592.1 |
| chr18_-_47018769 | 0.07 |
ENST00000583637.1
ENST00000578528.1 ENST00000578532.1 ENST00000580387.1 ENST00000579248.1 ENST00000581373.1 |
RPL17
|
ribosomal protein L17 |
| chr11_+_809647 | 0.07 |
ENST00000321153.4
|
RPLP2
|
ribosomal protein, large, P2 |
| chr15_-_64386120 | 0.07 |
ENST00000300030.3
|
FAM96A
|
family with sequence similarity 96, member A |
| chr17_-_27949911 | 0.07 |
ENST00000492276.2
ENST00000345068.5 ENST00000584602.1 |
CORO6
|
coronin 6 |
| chr12_-_102455902 | 0.07 |
ENST00000240079.6
|
CCDC53
|
coiled-coil domain containing 53 |
| chr7_+_116593536 | 0.07 |
ENST00000417919.1
|
ST7
|
suppression of tumorigenicity 7 |
| chr2_+_230787213 | 0.06 |
ENST00000409992.1
|
FBXO36
|
F-box protein 36 |
| chr4_-_1713977 | 0.06 |
ENST00000318386.4
|
SLBP
|
stem-loop binding protein |
| chr4_-_84406218 | 0.06 |
ENST00000515303.1
|
FAM175A
|
family with sequence similarity 175, member A |
| chr14_+_105992906 | 0.06 |
ENST00000392519.2
|
TMEM121
|
transmembrane protein 121 |
| chr2_-_63815628 | 0.06 |
ENST00000409562.3
|
WDPCP
|
WD repeat containing planar cell polarity effector |
| chr17_-_9479128 | 0.06 |
ENST00000574431.1
|
STX8
|
syntaxin 8 |
| chr1_-_24306835 | 0.06 |
ENST00000484146.2
|
SRSF10
|
serine/arginine-rich splicing factor 10 |
| chr11_-_73882029 | 0.06 |
ENST00000539061.1
|
C2CD3
|
C2 calcium-dependent domain containing 3 |
| chr3_-_157251383 | 0.06 |
ENST00000487753.1
ENST00000489602.1 ENST00000461299.1 ENST00000479987.1 |
VEPH1
|
ventricular zone expressed PH domain-containing 1 |
| chr16_+_11439286 | 0.06 |
ENST00000312499.5
ENST00000576027.1 |
RMI2
|
RecQ mediated genome instability 2 |
| chr6_+_135502501 | 0.06 |
ENST00000527615.1
ENST00000420123.2 ENST00000525369.1 ENST00000528774.1 ENST00000534121.1 ENST00000534044.1 ENST00000533624.1 |
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr2_-_148778258 | 0.06 |
ENST00000392857.5
ENST00000457954.1 ENST00000392858.1 ENST00000542387.1 |
ORC4
|
origin recognition complex, subunit 4 |
| chr1_-_32827682 | 0.06 |
ENST00000432622.1
|
FAM229A
|
family with sequence similarity 229, member A |
| chrX_+_24072833 | 0.06 |
ENST00000253039.4
|
EIF2S3
|
eukaryotic translation initiation factor 2, subunit 3 gamma, 52kDa |
| chr6_+_30034865 | 0.06 |
ENST00000376772.3
|
PPP1R11
|
protein phosphatase 1, regulatory (inhibitor) subunit 11 |
| chr1_+_150245177 | 0.06 |
ENST00000369098.3
|
C1orf54
|
chromosome 1 open reading frame 54 |
| chr3_+_19988566 | 0.06 |
ENST00000273047.4
|
RAB5A
|
RAB5A, member RAS oncogene family |
| chr3_+_69985792 | 0.06 |
ENST00000531774.1
|
MITF
|
microphthalmia-associated transcription factor |
| chr7_+_856246 | 0.06 |
ENST00000389574.3
ENST00000457378.2 ENST00000452783.2 ENST00000435699.1 ENST00000440380.1 ENST00000439679.1 ENST00000424128.1 |
SUN1
|
Sad1 and UNC84 domain containing 1 |
| chr19_+_47760777 | 0.06 |
ENST00000599398.1
ENST00000595659.1 |
CCDC9
|
coiled-coil domain containing 9 |
| chr6_-_90348440 | 0.06 |
ENST00000520441.1
ENST00000520318.1 ENST00000523377.1 |
LYRM2
|
LYR motif containing 2 |
| chr4_+_1714548 | 0.06 |
ENST00000605571.1
|
RP11-572O17.1
|
RP11-572O17.1 |
| chr18_-_47814032 | 0.06 |
ENST00000589548.1
ENST00000591474.1 |
CXXC1
|
CXXC finger protein 1 |
| chr15_-_64385981 | 0.06 |
ENST00000557835.1
ENST00000380290.3 ENST00000559950.1 |
FAM96A
|
family with sequence similarity 96, member A |
| chr15_-_98417780 | 0.05 |
ENST00000503874.3
|
LINC00923
|
long intergenic non-protein coding RNA 923 |
| chr19_+_49891475 | 0.05 |
ENST00000447857.3
|
CCDC155
|
coiled-coil domain containing 155 |
| chr2_+_10262857 | 0.05 |
ENST00000304567.5
|
RRM2
|
ribonucleotide reductase M2 |
| chr16_+_2732476 | 0.05 |
ENST00000301738.4
ENST00000564195.1 |
KCTD5
|
potassium channel tetramerization domain containing 5 |
| chr2_-_230787879 | 0.05 |
ENST00000435716.1
|
TRIP12
|
thyroid hormone receptor interactor 12 |
| chr1_-_150693318 | 0.05 |
ENST00000442853.1
ENST00000368995.4 ENST00000368993.2 ENST00000361824.2 ENST00000322343.7 |
HORMAD1
|
HORMA domain containing 1 |
| chr2_-_47403642 | 0.05 |
ENST00000456319.1
ENST00000409563.1 ENST00000272298.7 |
CALM2
|
calmodulin 2 (phosphorylase kinase, delta) |
| chr12_-_6772249 | 0.05 |
ENST00000467678.1
ENST00000493873.1 ENST00000423703.2 ENST00000412586.2 ENST00000444704.2 |
ING4
|
inhibitor of growth family, member 4 |
| chr18_-_47017956 | 0.05 |
ENST00000584895.1
ENST00000332968.6 ENST00000580210.1 ENST00000579408.1 |
RPL17-C18orf32
RPL17
|
RPL17-C18orf32 readthrough ribosomal protein L17 |
Network of associatons between targets according to the STRING database.
First level regulatory network of E2F8
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.1 | 0.3 | GO:0060829 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.1 | 0.2 | GO:1904899 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.1 | 0.2 | GO:0060629 | meiotic DNA double-strand break formation(GO:0042138) regulation of homologous chromosome segregation(GO:0060629) |
| 0.0 | 0.1 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.4 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:2000612 | thyroid-stimulating hormone secretion(GO:0070460) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.0 | 0.3 | GO:0019075 | virus maturation(GO:0019075) |
| 0.0 | 0.1 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 | 0.2 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.0 | 0.1 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) positive regulation of meiosis I(GO:0060903) |
| 0.0 | 0.1 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.0 | 0.2 | GO:0045349 | negative regulation of interferon-alpha production(GO:0032687) interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) |
| 0.0 | 0.2 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 0.0 | 0.1 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.0 | 0.2 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 0.1 | GO:1990426 | homologous recombination-dependent replication fork processing(GO:1990426) |
| 0.0 | 0.1 | GO:0032100 | positive regulation of response to food(GO:0032097) positive regulation of appetite(GO:0032100) |
| 0.0 | 0.2 | GO:0021997 | neural plate axis specification(GO:0021997) |
| 0.0 | 0.1 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.1 | GO:2000521 | negative regulation of immunological synapse formation(GO:2000521) |
| 0.0 | 0.1 | GO:2000657 | regulation of apolipoprotein binding(GO:2000656) negative regulation of apolipoprotein binding(GO:2000657) |
| 0.0 | 0.1 | GO:0003165 | Purkinje myocyte development(GO:0003165) |
| 0.0 | 0.1 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.0 | 0.1 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.0 | 0.4 | GO:0060044 | negative regulation of cardiac muscle cell proliferation(GO:0060044) |
| 0.0 | 0.2 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.2 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.3 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.1 | GO:0006051 | mannosamine metabolic process(GO:0006050) N-acetylmannosamine metabolic process(GO:0006051) |
| 0.0 | 0.0 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.0 | 0.1 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.0 | 0.1 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.0 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0000818 | nuclear MIS12/MIND complex(GO:0000818) |
| 0.0 | 0.1 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.1 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.1 | GO:0090661 | box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.1 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.0 | 0.2 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.1 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.1 | GO:0034448 | EGO complex(GO:0034448) |
| 0.0 | 0.1 | GO:0000931 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.0 | 0.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.0 | 0.1 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.0 | 0.3 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.1 | GO:0000224 | peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase activity(GO:0000224) |
| 0.0 | 0.1 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.1 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 0.0 | 0.2 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.1 | GO:0004996 | thyroid-stimulating hormone receptor activity(GO:0004996) |
| 0.0 | 0.2 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.0 | 0.1 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 0.2 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.4 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.1 | GO:0050610 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.0 | 0.1 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.1 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.0 | 0.1 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.2 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.2 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.3 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.1 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |