Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
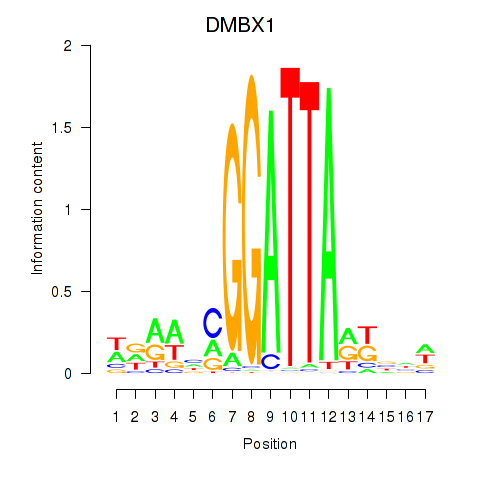
Results for DMBX1
Z-value: 0.99
Transcription factors associated with DMBX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
DMBX1
|
ENSG00000197587.6 | diencephalon/mesencephalon homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| DMBX1 | hg19_v2_chr1_+_46972668_46972669 | -0.99 | 6.6e-03 | Click! |
Activity profile of DMBX1 motif
Sorted Z-values of DMBX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_220252068 | 0.83 |
ENST00000430206.1
ENST00000429013.1 |
DNPEP
|
aspartyl aminopeptidase |
| chr17_+_4643337 | 0.75 |
ENST00000592813.1
|
ZMYND15
|
zinc finger, MYND-type containing 15 |
| chr5_+_71403280 | 0.67 |
ENST00000511641.2
|
MAP1B
|
microtubule-associated protein 1B |
| chr12_-_76461249 | 0.60 |
ENST00000551524.1
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr17_-_4643161 | 0.59 |
ENST00000574412.1
|
CXCL16
|
chemokine (C-X-C motif) ligand 16 |
| chr10_-_101491828 | 0.59 |
ENST00000370483.5
ENST00000016171.5 |
COX15
|
cytochrome c oxidase assembly homolog 15 (yeast) |
| chr1_-_143913143 | 0.56 |
ENST00000400889.1
|
FAM72D
|
family with sequence similarity 72, member D |
| chr14_+_50234309 | 0.54 |
ENST00000298307.5
|
KLHDC2
|
kelch domain containing 2 |
| chr1_+_81001398 | 0.53 |
ENST00000418041.1
ENST00000443104.1 |
RP5-887A10.1
|
RP5-887A10.1 |
| chr17_+_68100989 | 0.50 |
ENST00000585558.1
ENST00000392670.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr17_-_19651598 | 0.50 |
ENST00000570414.1
|
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr4_+_124571409 | 0.48 |
ENST00000514823.1
ENST00000511919.1 ENST00000508111.1 |
RP11-93L9.1
|
long intergenic non-protein coding RNA 1091 |
| chr15_-_60771280 | 0.46 |
ENST00000560072.1
ENST00000560406.1 ENST00000560520.1 ENST00000261520.4 ENST00000439632.1 |
NARG2
|
NMDA receptor regulated 2 |
| chr2_-_220252530 | 0.46 |
ENST00000521459.1
|
DNPEP
|
aspartyl aminopeptidase |
| chr6_+_10585979 | 0.45 |
ENST00000265012.4
|
GCNT2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr2_-_14541060 | 0.44 |
ENST00000418420.1
ENST00000417751.1 |
LINC00276
|
long intergenic non-protein coding RNA 276 |
| chr17_-_4643114 | 0.44 |
ENST00000293778.6
|
CXCL16
|
chemokine (C-X-C motif) ligand 16 |
| chr5_-_54988448 | 0.44 |
ENST00000503817.1
ENST00000512595.1 |
SLC38A9
|
solute carrier family 38, member 9 |
| chr15_+_93443419 | 0.41 |
ENST00000557381.1
ENST00000420239.2 |
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr17_+_4643300 | 0.40 |
ENST00000433935.1
|
ZMYND15
|
zinc finger, MYND-type containing 15 |
| chr3_+_136649311 | 0.39 |
ENST00000469404.1
ENST00000467911.1 |
NCK1
|
NCK adaptor protein 1 |
| chr6_-_33297013 | 0.38 |
ENST00000453407.1
|
DAXX
|
death-domain associated protein |
| chr1_+_35734562 | 0.37 |
ENST00000314607.6
ENST00000373297.2 |
ZMYM4
|
zinc finger, MYM-type 4 |
| chr22_+_31002990 | 0.37 |
ENST00000423350.1
|
TCN2
|
transcobalamin II |
| chr1_+_206138457 | 0.37 |
ENST00000367128.3
ENST00000431655.2 |
FAM72A
|
family with sequence similarity 72, member A |
| chr11_-_45940343 | 0.36 |
ENST00000532681.1
|
PEX16
|
peroxisomal biogenesis factor 16 |
| chr17_-_48207157 | 0.36 |
ENST00000330175.4
ENST00000503131.1 |
SAMD14
|
sterile alpha motif domain containing 14 |
| chr2_-_175711133 | 0.35 |
ENST00000409597.1
ENST00000413882.1 |
CHN1
|
chimerin 1 |
| chr7_-_135412925 | 0.35 |
ENST00000354042.4
|
SLC13A4
|
solute carrier family 13 (sodium/sulfate symporter), member 4 |
| chr6_-_111927062 | 0.34 |
ENST00000359831.4
|
TRAF3IP2
|
TRAF3 interacting protein 2 |
| chr3_-_121468513 | 0.32 |
ENST00000494517.1
ENST00000393667.3 |
GOLGB1
|
golgin B1 |
| chr8_-_125577940 | 0.30 |
ENST00000519168.1
ENST00000395508.2 |
MTSS1
|
metastasis suppressor 1 |
| chr3_-_121468602 | 0.30 |
ENST00000340645.5
|
GOLGB1
|
golgin B1 |
| chr4_+_41937131 | 0.28 |
ENST00000504986.1
ENST00000508448.1 ENST00000513702.1 ENST00000325094.5 |
TMEM33
|
transmembrane protein 33 |
| chr7_-_17500294 | 0.28 |
ENST00000439046.1
|
AC019117.2
|
AC019117.2 |
| chr17_-_19651654 | 0.28 |
ENST00000395555.3
|
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr8_-_110656995 | 0.26 |
ENST00000276646.9
ENST00000533065.1 |
SYBU
|
syntabulin (syntaxin-interacting) |
| chr1_+_120839005 | 0.25 |
ENST00000369390.3
ENST00000452190.1 |
FAM72B
|
family with sequence similarity 72, member B |
| chr17_+_68101117 | 0.25 |
ENST00000587698.1
ENST00000587892.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr17_-_19651668 | 0.25 |
ENST00000494157.2
ENST00000225740.6 |
ALDH3A1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr9_+_74920408 | 0.25 |
ENST00000451152.1
|
RP11-63P12.6
|
RP11-63P12.6 |
| chr6_+_89791507 | 0.24 |
ENST00000354922.3
|
PNRC1
|
proline-rich nuclear receptor coactivator 1 |
| chr10_+_74451883 | 0.24 |
ENST00000373053.3
ENST00000357157.6 |
MCU
|
mitochondrial calcium uniporter |
| chr9_+_74920335 | 0.23 |
ENST00000451596.2
ENST00000436054.1 |
RP11-63P12.6
|
RP11-63P12.6 |
| chr1_+_180875711 | 0.23 |
ENST00000434447.1
|
RP11-46A10.2
|
RP11-46A10.2 |
| chr6_+_130339710 | 0.22 |
ENST00000526087.1
ENST00000533560.1 ENST00000361794.2 |
L3MBTL3
|
l(3)mbt-like 3 (Drosophila) |
| chr4_-_54232144 | 0.22 |
ENST00000388940.4
ENST00000503450.1 ENST00000401642.3 |
SCFD2
|
sec1 family domain containing 2 |
| chr2_+_37571845 | 0.22 |
ENST00000537448.1
|
QPCT
|
glutaminyl-peptide cyclotransferase |
| chr12_-_49581152 | 0.21 |
ENST00000550811.1
|
TUBA1A
|
tubulin, alpha 1a |
| chr7_+_21582638 | 0.20 |
ENST00000409508.3
ENST00000328843.6 |
DNAH11
|
dynein, axonemal, heavy chain 11 |
| chr2_-_165812028 | 0.20 |
ENST00000303735.4
|
SLC38A11
|
solute carrier family 38, member 11 |
| chr12_-_110937351 | 0.20 |
ENST00000552130.2
|
VPS29
|
vacuolar protein sorting 29 homolog (S. cerevisiae) |
| chr18_+_52258390 | 0.17 |
ENST00000321600.1
|
DYNAP
|
dynactin associated protein |
| chr3_-_49058479 | 0.16 |
ENST00000440857.1
|
DALRD3
|
DALR anticodon binding domain containing 3 |
| chr11_+_61248583 | 0.16 |
ENST00000432063.2
ENST00000338608.2 |
PPP1R32
|
protein phosphatase 1, regulatory subunit 32 |
| chr12_-_56727676 | 0.15 |
ENST00000547572.1
ENST00000257931.5 ENST00000440411.3 |
PAN2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr3_-_19975665 | 0.15 |
ENST00000295824.9
ENST00000389256.4 |
EFHB
|
EF-hand domain family, member B |
| chr2_+_223725652 | 0.14 |
ENST00000357430.3
ENST00000392066.3 |
ACSL3
|
acyl-CoA synthetase long-chain family member 3 |
| chr15_+_63414760 | 0.14 |
ENST00000557972.1
|
LACTB
|
lactamase, beta |
| chr3_-_10362725 | 0.14 |
ENST00000397109.3
ENST00000428626.1 ENST00000445064.1 ENST00000431352.1 ENST00000397117.1 ENST00000337354.4 ENST00000383801.2 ENST00000432213.1 ENST00000350697.3 |
SEC13
|
SEC13 homolog (S. cerevisiae) |
| chr3_+_138340067 | 0.14 |
ENST00000479848.1
|
FAIM
|
Fas apoptotic inhibitory molecule |
| chr12_-_118628350 | 0.13 |
ENST00000537952.1
ENST00000537822.1 |
TAOK3
|
TAO kinase 3 |
| chr5_+_175740083 | 0.12 |
ENST00000332772.4
|
SIMC1
|
SUMO-interacting motifs containing 1 |
| chr3_+_99986036 | 0.12 |
ENST00000471098.1
|
TBC1D23
|
TBC1 domain family, member 23 |
| chr11_-_36619771 | 0.10 |
ENST00000311485.3
ENST00000527033.1 ENST00000532616.1 |
RAG2
|
recombination activating gene 2 |
| chr22_-_43411106 | 0.10 |
ENST00000453643.1
ENST00000263246.3 ENST00000337959.4 |
PACSIN2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr4_+_108815402 | 0.10 |
ENST00000503385.1
|
SGMS2
|
sphingomyelin synthase 2 |
| chr15_+_63413990 | 0.09 |
ENST00000261893.4
|
LACTB
|
lactamase, beta |
| chr16_-_31076332 | 0.08 |
ENST00000539836.3
ENST00000535577.1 ENST00000442862.2 |
ZNF668
|
zinc finger protein 668 |
| chr3_+_111393501 | 0.05 |
ENST00000393934.3
|
PLCXD2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr16_-_31076273 | 0.05 |
ENST00000426488.2
|
ZNF668
|
zinc finger protein 668 |
| chr8_+_86999516 | 0.04 |
ENST00000521564.1
|
ATP6V0D2
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2 |
| chr12_-_56321397 | 0.04 |
ENST00000557259.1
ENST00000549939.1 |
WIBG
|
within bgcn homolog (Drosophila) |
| chr17_-_48207115 | 0.04 |
ENST00000511964.1
|
SAMD14
|
sterile alpha motif domain containing 14 |
| chrX_-_72097698 | 0.03 |
ENST00000373530.1
|
DMRTC1
|
DMRT-like family C1 |
| chr21_+_18811205 | 0.03 |
ENST00000440664.1
|
C21orf37
|
chromosome 21 open reading frame 37 |
| chr2_-_27486951 | 0.03 |
ENST00000432351.1
|
SLC30A3
|
solute carrier family 30 (zinc transporter), member 3 |
| chr12_-_56727487 | 0.03 |
ENST00000548043.1
ENST00000425394.2 |
PAN2
|
PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr4_+_619386 | 0.03 |
ENST00000496514.1
|
PDE6B
|
phosphodiesterase 6B, cGMP-specific, rod, beta |
| chr1_+_12851545 | 0.02 |
ENST00000332296.7
|
PRAMEF1
|
PRAME family member 1 |
| chr2_-_70780572 | 0.02 |
ENST00000450929.1
|
TGFA
|
transforming growth factor, alpha |
| chrX_+_72062802 | 0.02 |
ENST00000373533.1
|
DMRTC1B
|
DMRT-like family C1B |
| chr10_+_86004802 | 0.01 |
ENST00000359452.4
ENST00000358110.5 ENST00000372092.3 |
RGR
|
retinal G protein coupled receptor |
| chr4_+_619347 | 0.01 |
ENST00000255622.6
|
PDE6B
|
phosphodiesterase 6B, cGMP-specific, rod, beta |
| chr3_-_149293990 | 0.01 |
ENST00000472417.1
|
WWTR1
|
WW domain containing transcription regulator 1 |
| chr4_-_170679024 | 0.01 |
ENST00000393381.2
|
C4orf27
|
chromosome 4 open reading frame 27 |
Network of associatons between targets according to the STRING database.
First level regulatory network of DMBX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:1903676 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.1 | 0.4 | GO:0072738 | response to diamide(GO:0072737) cellular response to diamide(GO:0072738) |
| 0.1 | 0.4 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 0.1 | 0.3 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.1 | 0.2 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.1 | 0.5 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 1.0 | GO:0010818 | T cell chemotaxis(GO:0010818) |
| 0.0 | 0.3 | GO:0030035 | microspike assembly(GO:0030035) epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.4 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.7 | GO:0061162 | establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.0 | 0.2 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.6 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.2 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.3 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.2 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.2 | GO:0003356 | regulation of cilium beat frequency(GO:0003356) |
| 0.0 | 0.8 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.3 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.2 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.6 | GO:0070069 | cytochrome complex(GO:0070069) |
| 0.0 | 0.3 | GO:0097433 | dense body(GO:0097433) |
| 0.0 | 0.4 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.1 | 0.4 | GO:0071074 | eukaryotic initiation factor eIF2 binding(GO:0071074) |
| 0.1 | 0.2 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.1 | 0.5 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 1.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.8 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.4 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.0 | 1.0 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.2 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.6 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.3 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.4 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.0 | 0.1 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.8 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.4 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |