Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
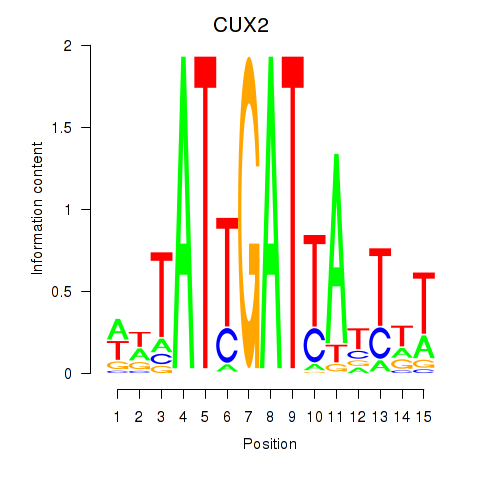
Results for CUX2
Z-value: 0.67
Transcription factors associated with CUX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CUX2
|
ENSG00000111249.9 | cut like homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CUX2 | hg19_v2_chr12_+_111471828_111471975 | -0.64 | 3.6e-01 | Click! |
Activity profile of CUX2 motif
Sorted Z-values of CUX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_92777606 | 0.65 |
ENST00000437805.1
ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L
|
sterile alpha motif domain containing 9-like |
| chr21_+_42792442 | 0.59 |
ENST00000398600.2
|
MX1
|
myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) |
| chrX_-_80457385 | 0.38 |
ENST00000451455.1
ENST00000436386.1 ENST00000358130.2 |
HMGN5
|
high mobility group nucleosome binding domain 5 |
| chr1_-_109618566 | 0.34 |
ENST00000338366.5
|
TAF13
|
TAF13 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 18kDa |
| chr6_+_26273144 | 0.32 |
ENST00000377733.2
|
HIST1H2BI
|
histone cluster 1, H2bi |
| chr4_+_69962212 | 0.32 |
ENST00000508661.1
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr5_+_40841410 | 0.28 |
ENST00000381677.3
|
CARD6
|
caspase recruitment domain family, member 6 |
| chr12_-_11339543 | 0.28 |
ENST00000334266.1
|
TAS2R42
|
taste receptor, type 2, member 42 |
| chr12_+_28605426 | 0.27 |
ENST00000542801.1
|
CCDC91
|
coiled-coil domain containing 91 |
| chr12_-_3982511 | 0.27 |
ENST00000427057.2
ENST00000228820.4 |
PARP11
|
poly (ADP-ribose) polymerase family, member 11 |
| chr5_+_115358088 | 0.24 |
ENST00000600981.3
|
CTD-2287O16.3
|
CTD-2287O16.3 |
| chrX_+_33744503 | 0.23 |
ENST00000439992.1
|
RP11-305F18.1
|
RP11-305F18.1 |
| chr6_+_31105426 | 0.23 |
ENST00000547221.1
|
PSORS1C1
|
psoriasis susceptibility 1 candidate 1 |
| chr12_+_6554021 | 0.23 |
ENST00000266557.3
|
CD27
|
CD27 molecule |
| chr12_-_106477805 | 0.23 |
ENST00000553094.1
ENST00000549704.1 |
NUAK1
|
NUAK family, SNF1-like kinase, 1 |
| chrM_+_4431 | 0.23 |
ENST00000361453.3
|
MT-ND2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr16_-_30621663 | 0.22 |
ENST00000287461.3
|
ZNF689
|
zinc finger protein 689 |
| chr4_+_70916119 | 0.22 |
ENST00000246896.3
ENST00000511674.1 |
HTN1
|
histatin 1 |
| chr20_-_44516256 | 0.22 |
ENST00000372519.3
|
SPATA25
|
spermatogenesis associated 25 |
| chr5_-_54988559 | 0.22 |
ENST00000502247.1
|
SLC38A9
|
solute carrier family 38, member 9 |
| chr3_+_157827841 | 0.21 |
ENST00000295930.3
ENST00000471994.1 ENST00000464171.1 ENST00000312179.6 ENST00000475278.2 |
RSRC1
|
arginine/serine-rich coiled-coil 1 |
| chr3_+_172034218 | 0.20 |
ENST00000366261.2
|
AC092964.1
|
Uncharacterized protein |
| chr8_-_8318847 | 0.20 |
ENST00000521218.1
|
CTA-398F10.2
|
CTA-398F10.2 |
| chr14_+_50159813 | 0.20 |
ENST00000359332.2
ENST00000553274.1 ENST00000557128.1 |
KLHDC1
|
kelch domain containing 1 |
| chr12_-_10601963 | 0.20 |
ENST00000543893.1
|
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1 |
| chr19_+_18726786 | 0.20 |
ENST00000594709.1
|
TMEM59L
|
transmembrane protein 59-like |
| chr15_-_83837983 | 0.19 |
ENST00000562702.1
|
HDGFRP3
|
Hepatoma-derived growth factor-related protein 3 |
| chr12_-_68797580 | 0.19 |
ENST00000539404.1
|
RP11-81H14.2
|
RP11-81H14.2 |
| chr1_+_158985457 | 0.18 |
ENST00000567661.1
ENST00000474473.1 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr11_+_57365150 | 0.18 |
ENST00000457869.1
ENST00000340687.6 ENST00000378323.4 ENST00000378324.2 ENST00000403558.1 |
SERPING1
|
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr19_-_4535233 | 0.18 |
ENST00000381848.3
ENST00000588887.1 ENST00000586133.1 |
PLIN5
|
perilipin 5 |
| chr1_-_23340400 | 0.18 |
ENST00000440767.2
|
C1orf234
|
chromosome 1 open reading frame 234 |
| chr11_-_77734260 | 0.18 |
ENST00000353172.5
|
KCTD14
|
potassium channel tetramerization domain containing 14 |
| chr17_+_68047418 | 0.18 |
ENST00000586373.1
ENST00000588782.1 |
LINC01028
|
long intergenic non-protein coding RNA 1028 |
| chr6_+_30035307 | 0.18 |
ENST00000376765.2
ENST00000376763.1 |
PPP1R11
|
protein phosphatase 1, regulatory (inhibitor) subunit 11 |
| chr15_+_42697065 | 0.18 |
ENST00000565559.1
|
CAPN3
|
calpain 3, (p94) |
| chr7_+_143771275 | 0.18 |
ENST00000408898.2
|
OR2A25
|
olfactory receptor, family 2, subfamily A, member 25 |
| chr11_+_1092184 | 0.17 |
ENST00000361558.6
|
MUC2
|
mucin 2, oligomeric mucus/gel-forming |
| chr3_-_194072019 | 0.17 |
ENST00000429275.1
ENST00000323830.3 |
CPN2
|
carboxypeptidase N, polypeptide 2 |
| chr11_-_47736896 | 0.17 |
ENST00000525123.1
ENST00000528244.1 ENST00000532595.1 ENST00000529154.1 ENST00000530969.1 |
AGBL2
|
ATP/GTP binding protein-like 2 |
| chr18_-_51750948 | 0.17 |
ENST00000583046.1
ENST00000398398.2 |
MBD2
|
methyl-CpG binding domain protein 2 |
| chr21_-_39705286 | 0.16 |
ENST00000414189.1
|
AP001422.3
|
AP001422.3 |
| chr1_+_104159999 | 0.16 |
ENST00000414303.2
ENST00000423678.1 |
AMY2A
|
amylase, alpha 2A (pancreatic) |
| chr10_+_5238793 | 0.16 |
ENST00000263126.1
|
AKR1C4
|
aldo-keto reductase family 1, member C4 |
| chr5_-_135290651 | 0.16 |
ENST00000522943.1
ENST00000514447.2 |
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr12_-_91546926 | 0.16 |
ENST00000550758.1
|
DCN
|
decorin |
| chr3_+_98699880 | 0.15 |
ENST00000473756.1
|
LINC00973
|
long intergenic non-protein coding RNA 973 |
| chr15_-_38519066 | 0.15 |
ENST00000561320.1
ENST00000561161.1 |
RP11-346D14.1
|
RP11-346D14.1 |
| chr4_+_646960 | 0.15 |
ENST00000488061.1
ENST00000429163.2 |
PDE6B
|
phosphodiesterase 6B, cGMP-specific, rod, beta |
| chr1_+_67632083 | 0.15 |
ENST00000347310.5
ENST00000371002.1 |
IL23R
|
interleukin 23 receptor |
| chr5_-_55412774 | 0.15 |
ENST00000434982.2
|
ANKRD55
|
ankyrin repeat domain 55 |
| chr1_-_151813033 | 0.15 |
ENST00000454109.1
|
C2CD4D
|
C2 calcium-dependent domain containing 4D |
| chr4_-_110624564 | 0.15 |
ENST00000352981.3
ENST00000265164.2 ENST00000505486.1 |
CASP6
|
caspase 6, apoptosis-related cysteine peptidase |
| chr5_+_133706865 | 0.14 |
ENST00000265339.2
|
UBE2B
|
ubiquitin-conjugating enzyme E2B |
| chr8_-_93978309 | 0.14 |
ENST00000517858.1
ENST00000378861.5 |
TRIQK
|
triple QxxK/R motif containing |
| chr19_-_58864848 | 0.14 |
ENST00000263100.3
|
A1BG
|
alpha-1-B glycoprotein |
| chr11_+_110225855 | 0.14 |
ENST00000526605.1
ENST00000526703.1 |
RP11-347E10.1
|
RP11-347E10.1 |
| chr1_+_44514040 | 0.14 |
ENST00000431800.1
ENST00000437643.1 |
RP5-1198O20.4
|
RP5-1198O20.4 |
| chr13_+_21714653 | 0.14 |
ENST00000382533.4
|
SAP18
|
Sin3A-associated protein, 18kDa |
| chr1_+_234765057 | 0.14 |
ENST00000429269.1
|
LINC00184
|
long intergenic non-protein coding RNA 184 |
| chr17_-_34890617 | 0.13 |
ENST00000586886.1
ENST00000585719.1 ENST00000585818.1 |
MYO19
|
myosin XIX |
| chr19_+_507299 | 0.13 |
ENST00000359315.5
|
TPGS1
|
tubulin polyglutamylase complex subunit 1 |
| chr17_-_54893250 | 0.13 |
ENST00000397862.2
|
C17orf67
|
chromosome 17 open reading frame 67 |
| chr16_-_28192360 | 0.13 |
ENST00000570033.1
|
XPO6
|
exportin 6 |
| chr3_+_138066539 | 0.13 |
ENST00000289104.4
|
MRAS
|
muscle RAS oncogene homolog |
| chr9_-_100881466 | 0.13 |
ENST00000341469.2
ENST00000342043.3 ENST00000375098.3 |
TRIM14
|
tripartite motif containing 14 |
| chr14_-_73997901 | 0.13 |
ENST00000557603.1
ENST00000556455.1 |
HEATR4
|
HEAT repeat containing 4 |
| chr16_-_740354 | 0.13 |
ENST00000293883.4
|
WDR24
|
WD repeat domain 24 |
| chr21_-_28215332 | 0.13 |
ENST00000517777.1
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr4_-_90756769 | 0.13 |
ENST00000345009.4
ENST00000505199.1 ENST00000502987.1 |
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chrM_+_10758 | 0.13 |
ENST00000361381.2
|
MT-ND4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr17_-_56084578 | 0.12 |
ENST00000582730.2
ENST00000584773.1 ENST00000585096.1 ENST00000258962.4 |
SRSF1
|
serine/arginine-rich splicing factor 1 |
| chr6_+_46761118 | 0.12 |
ENST00000230588.4
|
MEP1A
|
meprin A, alpha (PABA peptide hydrolase) |
| chr19_-_37697976 | 0.12 |
ENST00000588873.1
|
CTC-454I21.3
|
Uncharacterized protein; Zinc finger protein 585B |
| chr2_+_131769256 | 0.12 |
ENST00000355771.3
|
ARHGEF4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr7_+_120629653 | 0.12 |
ENST00000450913.2
ENST00000340646.5 |
CPED1
|
cadherin-like and PC-esterase domain containing 1 |
| chr6_+_24667257 | 0.12 |
ENST00000537591.1
ENST00000230048.4 |
ACOT13
|
acyl-CoA thioesterase 13 |
| chr19_+_21106081 | 0.12 |
ENST00000300540.3
ENST00000595854.1 ENST00000601284.1 ENST00000328178.8 ENST00000599885.1 ENST00000596476.1 ENST00000345030.6 |
ZNF85
|
zinc finger protein 85 |
| chr6_+_127898312 | 0.12 |
ENST00000329722.7
|
C6orf58
|
chromosome 6 open reading frame 58 |
| chr2_-_99870744 | 0.12 |
ENST00000409238.1
ENST00000423800.1 |
LYG2
|
lysozyme G-like 2 |
| chr6_-_146057144 | 0.12 |
ENST00000367519.3
|
EPM2A
|
epilepsy, progressive myoclonus type 2A, Lafora disease (laforin) |
| chr1_+_149239529 | 0.12 |
ENST00000457216.2
|
RP11-403I13.4
|
RP11-403I13.4 |
| chr1_+_161035655 | 0.12 |
ENST00000600454.1
|
AL591806.1
|
Uncharacterized protein |
| chr9_+_131683174 | 0.12 |
ENST00000372592.3
ENST00000428610.1 |
PHYHD1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr2_-_214017151 | 0.12 |
ENST00000452786.1
|
IKZF2
|
IKAROS family zinc finger 2 (Helios) |
| chr5_-_33297946 | 0.11 |
ENST00000510327.1
|
CTD-2066L21.3
|
CTD-2066L21.3 |
| chr5_-_135290705 | 0.11 |
ENST00000274507.1
|
LECT2
|
leukocyte cell-derived chemotaxin 2 |
| chr17_+_67498295 | 0.11 |
ENST00000589295.1
|
MAP2K6
|
mitogen-activated protein kinase kinase 6 |
| chr3_-_180707306 | 0.11 |
ENST00000479269.1
|
DNAJC19
|
DnaJ (Hsp40) homolog, subfamily C, member 19 |
| chrX_+_36246735 | 0.11 |
ENST00000378653.3
|
CXorf30
|
chromosome X open reading frame 30 |
| chr19_+_49588677 | 0.11 |
ENST00000598984.1
ENST00000598441.1 |
SNRNP70
|
small nuclear ribonucleoprotein 70kDa (U1) |
| chr6_-_27799305 | 0.11 |
ENST00000357549.2
|
HIST1H4K
|
histone cluster 1, H4k |
| chr6_+_146056706 | 0.11 |
ENST00000603994.1
|
RP3-466P17.1
|
RP3-466P17.1 |
| chr6_+_27833034 | 0.11 |
ENST00000357320.2
|
HIST1H2AL
|
histone cluster 1, H2al |
| chr4_+_69962185 | 0.11 |
ENST00000305231.7
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr14_+_88490894 | 0.11 |
ENST00000556033.1
ENST00000553929.1 ENST00000555996.1 ENST00000556673.1 ENST00000557339.1 ENST00000556684.1 |
RP11-300J18.3
|
long intergenic non-protein coding RNA 1146 |
| chr19_+_49588690 | 0.11 |
ENST00000221448.5
|
SNRNP70
|
small nuclear ribonucleoprotein 70kDa (U1) |
| chr2_+_232573208 | 0.11 |
ENST00000409115.3
|
PTMA
|
prothymosin, alpha |
| chr2_+_170655322 | 0.11 |
ENST00000260956.4
ENST00000417292.1 |
SSB
|
Sjogren syndrome antigen B (autoantigen La) |
| chr9_-_27297126 | 0.10 |
ENST00000380031.1
ENST00000537675.1 ENST00000380032.3 |
EQTN
|
equatorin, sperm acrosome associated |
| chr4_-_103746683 | 0.10 |
ENST00000504211.1
ENST00000508476.1 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr17_+_41150290 | 0.10 |
ENST00000589037.1
ENST00000253788.5 |
RPL27
|
ribosomal protein L27 |
| chr16_-_75467318 | 0.10 |
ENST00000283882.3
|
CFDP1
|
craniofacial development protein 1 |
| chr6_-_32160622 | 0.10 |
ENST00000487761.1
ENST00000375040.3 |
GPSM3
|
G-protein signaling modulator 3 |
| chrX_-_44402231 | 0.10 |
ENST00000378045.4
|
FUNDC1
|
FUN14 domain containing 1 |
| chr5_-_133706695 | 0.10 |
ENST00000521755.1
ENST00000523054.1 ENST00000435240.2 ENST00000609654.1 ENST00000536186.1 ENST00000609383.1 |
CDKL3
|
cyclin-dependent kinase-like 3 |
| chr10_-_4720301 | 0.10 |
ENST00000449712.1
|
LINC00704
|
long intergenic non-protein coding RNA 704 |
| chr22_-_26961328 | 0.10 |
ENST00000398110.2
|
TPST2
|
tyrosylprotein sulfotransferase 2 |
| chr9_-_130497565 | 0.10 |
ENST00000336067.6
ENST00000373281.5 ENST00000373284.5 ENST00000458505.3 |
TOR2A
|
torsin family 2, member A |
| chr1_-_89736434 | 0.10 |
ENST00000370459.3
|
GBP5
|
guanylate binding protein 5 |
| chr13_-_86373536 | 0.10 |
ENST00000400286.2
|
SLITRK6
|
SLIT and NTRK-like family, member 6 |
| chr2_+_27886330 | 0.10 |
ENST00000326019.6
|
SLC4A1AP
|
solute carrier family 4 (anion exchanger), member 1, adaptor protein |
| chr2_+_172309634 | 0.10 |
ENST00000339506.3
|
DCAF17
|
DDB1 and CUL4 associated factor 17 |
| chr4_+_74275057 | 0.10 |
ENST00000511370.1
|
ALB
|
albumin |
| chr19_+_54466179 | 0.10 |
ENST00000270458.2
|
CACNG8
|
calcium channel, voltage-dependent, gamma subunit 8 |
| chr7_-_23510086 | 0.09 |
ENST00000258729.3
|
IGF2BP3
|
insulin-like growth factor 2 mRNA binding protein 3 |
| chr8_-_82598511 | 0.09 |
ENST00000449740.2
ENST00000311489.4 ENST00000521360.1 ENST00000519964.1 ENST00000518202.1 |
IMPA1
|
inositol(myo)-1(or 4)-monophosphatase 1 |
| chr2_+_86333340 | 0.09 |
ENST00000409783.2
ENST00000409277.3 |
PTCD3
|
pentatricopeptide repeat domain 3 |
| chr11_-_59633951 | 0.09 |
ENST00000257264.3
|
TCN1
|
transcobalamin I (vitamin B12 binding protein, R binder family) |
| chr4_+_71587669 | 0.09 |
ENST00000381006.3
ENST00000226328.4 |
RUFY3
|
RUN and FYVE domain containing 3 |
| chr12_-_6579808 | 0.09 |
ENST00000535180.1
ENST00000400911.3 |
VAMP1
|
vesicle-associated membrane protein 1 (synaptobrevin 1) |
| chr4_-_103747011 | 0.09 |
ENST00000350435.7
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr3_+_136676707 | 0.09 |
ENST00000329582.4
|
IL20RB
|
interleukin 20 receptor beta |
| chr9_+_74729511 | 0.09 |
ENST00000545168.1
|
GDA
|
guanine deaminase |
| chr7_+_23338819 | 0.09 |
ENST00000466681.1
|
MALSU1
|
mitochondrial assembly of ribosomal large subunit 1 |
| chr6_+_131958436 | 0.09 |
ENST00000357639.3
ENST00000543135.1 ENST00000427148.2 ENST00000358229.5 |
ENPP3
|
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr13_-_36920420 | 0.09 |
ENST00000438666.2
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome) |
| chr1_+_64014588 | 0.09 |
ENST00000371086.2
ENST00000340052.3 |
DLEU2L
|
deleted in lymphocytic leukemia 2-like |
| chr4_-_103746924 | 0.09 |
ENST00000505207.1
ENST00000502404.1 ENST00000507845.1 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr8_+_36641842 | 0.09 |
ENST00000523973.1
ENST00000399881.3 |
KCNU1
|
potassium channel, subfamily U, member 1 |
| chr2_+_149447783 | 0.09 |
ENST00000449013.1
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr15_-_99791413 | 0.09 |
ENST00000394129.2
ENST00000558663.1 ENST00000394135.3 ENST00000561365.1 ENST00000560279.1 |
TTC23
|
tetratricopeptide repeat domain 23 |
| chr1_+_202385953 | 0.09 |
ENST00000466968.1
|
PPP1R12B
|
protein phosphatase 1, regulatory subunit 12B |
| chr19_+_42301079 | 0.09 |
ENST00000596544.1
|
CEACAM3
|
carcinoembryonic antigen-related cell adhesion molecule 3 |
| chr19_-_52674896 | 0.09 |
ENST00000322146.8
ENST00000597065.1 |
ZNF836
|
zinc finger protein 836 |
| chr1_-_35450897 | 0.09 |
ENST00000373337.3
|
ZMYM6NB
|
ZMYM6 neighbor |
| chr5_+_179135246 | 0.09 |
ENST00000508787.1
|
CANX
|
calnexin |
| chr9_-_106087316 | 0.09 |
ENST00000411575.1
|
RP11-341A22.2
|
RP11-341A22.2 |
| chr10_+_99627889 | 0.09 |
ENST00000596005.1
|
GOLGA7B
|
Golgin subfamily A member 7B; cDNA FLJ43465 fis, clone OCBBF2036476 |
| chr6_+_168434678 | 0.09 |
ENST00000496008.1
|
KIF25
|
kinesin family member 25 |
| chr17_-_6524159 | 0.09 |
ENST00000589033.1
|
KIAA0753
|
KIAA0753 |
| chr21_+_17214724 | 0.09 |
ENST00000449491.1
|
USP25
|
ubiquitin specific peptidase 25 |
| chr4_-_69083720 | 0.08 |
ENST00000432593.3
|
TMPRSS11BNL
|
TMPRSS11B N-terminal like |
| chr16_-_740419 | 0.08 |
ENST00000248142.6
|
WDR24
|
WD repeat domain 24 |
| chr3_+_157828152 | 0.08 |
ENST00000476899.1
|
RSRC1
|
arginine/serine-rich coiled-coil 1 |
| chr17_-_80797886 | 0.08 |
ENST00000572562.1
|
ZNF750
|
zinc finger protein 750 |
| chr1_-_243417762 | 0.08 |
ENST00000522191.1
|
CEP170
|
centrosomal protein 170kDa |
| chr19_+_58570605 | 0.08 |
ENST00000359978.6
ENST00000401053.4 ENST00000439855.2 ENST00000313434.5 ENST00000511556.1 ENST00000506786.1 |
ZNF135
|
zinc finger protein 135 |
| chrX_-_47489244 | 0.08 |
ENST00000469388.1
ENST00000396992.3 ENST00000377005.2 |
CFP
|
complement factor properdin |
| chr2_+_58655461 | 0.08 |
ENST00000429095.1
ENST00000429664.1 ENST00000452840.1 |
AC007092.1
|
long intergenic non-protein coding RNA 1122 |
| chr4_+_95128996 | 0.08 |
ENST00000457823.2
|
SMARCAD1
|
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
| chr2_+_169926047 | 0.08 |
ENST00000428522.1
ENST00000450153.1 ENST00000421653.1 |
DHRS9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr22_-_36556821 | 0.08 |
ENST00000531095.1
ENST00000397293.2 ENST00000349314.2 |
APOL3
|
apolipoprotein L, 3 |
| chrX_+_10124977 | 0.08 |
ENST00000380833.4
|
CLCN4
|
chloride channel, voltage-sensitive 4 |
| chr12_+_123944070 | 0.08 |
ENST00000412157.2
|
SNRNP35
|
small nuclear ribonucleoprotein 35kDa (U11/U12) |
| chr2_+_101591314 | 0.08 |
ENST00000450763.1
|
NPAS2
|
neuronal PAS domain protein 2 |
| chr1_+_12976450 | 0.08 |
ENST00000361079.2
|
PRAMEF7
|
PRAME family member 7 |
| chr3_-_48598547 | 0.08 |
ENST00000536104.1
ENST00000452531.1 |
PFKFB4
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 |
| chr22_+_24198890 | 0.08 |
ENST00000345044.6
|
SLC2A11
|
solute carrier family 2 (facilitated glucose transporter), member 11 |
| chr14_+_67831576 | 0.08 |
ENST00000555876.1
|
EIF2S1
|
eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa |
| chr1_-_89591749 | 0.08 |
ENST00000370466.3
|
GBP2
|
guanylate binding protein 2, interferon-inducible |
| chr22_+_20703829 | 0.08 |
ENST00000583722.1
|
FAM230A
|
family with sequence similarity 230, member A |
| chr1_+_104293028 | 0.08 |
ENST00000370079.3
|
AMY1C
|
amylase, alpha 1C (salivary) |
| chr14_-_81425828 | 0.08 |
ENST00000555529.1
ENST00000556042.1 ENST00000556981.1 |
CEP128
|
centrosomal protein 128kDa |
| chr8_+_54764346 | 0.08 |
ENST00000297313.3
ENST00000344277.6 |
RGS20
|
regulator of G-protein signaling 20 |
| chr6_-_138539627 | 0.08 |
ENST00000527246.2
|
PBOV1
|
prostate and breast cancer overexpressed 1 |
| chr7_-_91509986 | 0.08 |
ENST00000456229.1
ENST00000442961.1 ENST00000406735.2 ENST00000419292.1 ENST00000351870.3 |
MTERF
|
mitochondrial transcription termination factor |
| chr4_+_159131596 | 0.08 |
ENST00000512481.1
|
TMEM144
|
transmembrane protein 144 |
| chr19_+_58193388 | 0.08 |
ENST00000596085.1
ENST00000594684.1 |
ZNF551
AC003006.7
|
zinc finger protein 551 Uncharacterized protein |
| chr20_+_31755934 | 0.08 |
ENST00000354932.5
|
BPIFA2
|
BPI fold containing family A, member 2 |
| chr12_+_21168630 | 0.08 |
ENST00000421593.2
|
SLCO1B7
|
solute carrier organic anion transporter family, member 1B7 (non-functional) |
| chr3_+_44626446 | 0.07 |
ENST00000441021.1
ENST00000322734.2 |
ZNF660
|
zinc finger protein 660 |
| chr11_+_43380459 | 0.07 |
ENST00000299240.6
ENST00000039989.4 |
TTC17
|
tetratricopeptide repeat domain 17 |
| chr20_+_44330651 | 0.07 |
ENST00000305479.2
|
WFDC13
|
WAP four-disulfide core domain 13 |
| chr5_+_1801503 | 0.07 |
ENST00000274137.5
ENST00000469176.1 |
NDUFS6
|
NADH dehydrogenase (ubiquinone) Fe-S protein 6, 13kDa (NADH-coenzyme Q reductase) |
| chr9_+_134001455 | 0.07 |
ENST00000531584.1
|
NUP214
|
nucleoporin 214kDa |
| chr5_+_173472607 | 0.07 |
ENST00000303177.3
ENST00000519867.1 |
NSG2
|
Neuron-specific protein family member 2 |
| chr12_+_128399965 | 0.07 |
ENST00000540882.1
ENST00000542089.1 |
LINC00507
|
long intergenic non-protein coding RNA 507 |
| chr8_+_125954281 | 0.07 |
ENST00000510897.2
ENST00000533286.1 |
LINC00964
|
long intergenic non-protein coding RNA 964 |
| chr19_+_35417939 | 0.07 |
ENST00000601142.1
ENST00000426813.2 |
ZNF30
|
zinc finger protein 30 |
| chr5_+_133707252 | 0.07 |
ENST00000506787.1
ENST00000507277.1 |
UBE2B
|
ubiquitin-conjugating enzyme E2B |
| chr2_+_232573222 | 0.07 |
ENST00000341369.7
ENST00000409683.1 |
PTMA
|
prothymosin, alpha |
| chr1_-_153949751 | 0.07 |
ENST00000428469.1
|
JTB
|
jumping translocation breakpoint |
| chr6_+_56911336 | 0.07 |
ENST00000370733.4
|
KIAA1586
|
KIAA1586 |
| chr15_+_58724184 | 0.07 |
ENST00000433326.2
|
LIPC
|
lipase, hepatic |
| chr1_+_8378140 | 0.07 |
ENST00000377479.2
|
SLC45A1
|
solute carrier family 45, member 1 |
| chr5_-_74326724 | 0.07 |
ENST00000322348.4
|
GCNT4
|
glucosaminyl (N-acetyl) transferase 4, core 2 |
| chr12_+_15475331 | 0.07 |
ENST00000281171.4
|
PTPRO
|
protein tyrosine phosphatase, receptor type, O |
| chr1_-_19615744 | 0.07 |
ENST00000361640.4
|
AKR7A3
|
aldo-keto reductase family 7, member A3 (aflatoxin aldehyde reductase) |
| chr15_-_51397473 | 0.07 |
ENST00000327536.5
|
TNFAIP8L3
|
tumor necrosis factor, alpha-induced protein 8-like 3 |
| chr1_-_110933663 | 0.07 |
ENST00000369781.4
ENST00000541986.1 ENST00000369779.4 |
SLC16A4
|
solute carrier family 16, member 4 |
| chr2_+_9778872 | 0.07 |
ENST00000478468.1
|
RP11-521D12.1
|
RP11-521D12.1 |
| chr11_+_35201826 | 0.07 |
ENST00000531873.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr16_-_66583994 | 0.07 |
ENST00000564917.1
|
TK2
|
thymidine kinase 2, mitochondrial |
| chr14_-_24020858 | 0.07 |
ENST00000419474.3
|
ZFHX2
|
zinc finger homeobox 2 |
| chr3_+_197677379 | 0.06 |
ENST00000442341.1
|
RPL35A
|
ribosomal protein L35a |
| chr5_-_43397184 | 0.06 |
ENST00000513525.1
|
CCL28
|
chemokine (C-C motif) ligand 28 |
| chr6_-_131321863 | 0.06 |
ENST00000528282.1
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr5_+_140593509 | 0.06 |
ENST00000341948.4
|
PCDHB13
|
protocadherin beta 13 |
| chr8_+_104892639 | 0.06 |
ENST00000436393.2
|
RIMS2
|
regulating synaptic membrane exocytosis 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of CUX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.1 | 0.2 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) negative regulation of histone phosphorylation(GO:0033128) positive regulation of meiosis I(GO:0060903) |
| 0.0 | 0.2 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.7 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.3 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.0 | 0.1 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.0 | 0.2 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.1 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 0.1 | GO:0072616 | interleukin-18 secretion(GO:0072616) |
| 0.0 | 0.1 | GO:0001897 | cytolysis by symbiont of host cells(GO:0001897) |
| 0.0 | 0.2 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.2 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.0 | 0.1 | GO:0006147 | guanine catabolic process(GO:0006147) |
| 0.0 | 0.6 | GO:0003373 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.2 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.0 | 0.1 | GO:0046603 | negative regulation of mitotic centrosome separation(GO:0046603) |
| 0.0 | 0.2 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.1 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.1 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.1 | GO:0002121 | inter-male aggressive behavior(GO:0002121) |
| 0.0 | 0.1 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.1 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.0 | 0.1 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.0 | 0.1 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.0 | 0.0 | GO:1901896 | positive regulation of calcium-transporting ATPase activity(GO:1901896) |
| 0.0 | 0.0 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.0 | 0.2 | GO:0009753 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.0 | 0.1 | GO:0033037 | polysaccharide localization(GO:0033037) |
| 0.0 | 0.1 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.2 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.2 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 0.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.3 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.1 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.0 | 0.1 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.1 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.4 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.0 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) ornithine transport(GO:0015822) |
| 0.0 | 0.0 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 0.0 | 0.0 | GO:0060490 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.0 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.1 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.0 | 0.1 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.0 | GO:0051389 | inactivation of MAPKK activity(GO:0051389) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.0 | 0.0 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 0.1 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.0 | 0.0 | GO:2000646 | lipid transport involved in lipid storage(GO:0010877) NMDA glutamate receptor clustering(GO:0097114) positive regulation of receptor catabolic process(GO:2000646) |
| 0.0 | 0.1 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.1 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.4 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.0 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.0 | 0.1 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.0 | 0.1 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.1 | GO:0045959 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0070703 | inner mucus layer(GO:0070702) outer mucus layer(GO:0070703) |
| 0.0 | 0.2 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 0.1 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.0 | 0.1 | GO:0043614 | multi-eIF complex(GO:0043614) translation preinitiation complex(GO:0070993) astrocyte end-foot(GO:0097450) |
| 0.0 | 0.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.2 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.3 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.1 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 0.0 | 0.0 | GO:0097635 | extrinsic component of autophagosome membrane(GO:0097635) |
| 0.0 | 0.2 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.0 | 0.1 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.0 | GO:0098844 | postsynaptic endocytic zone(GO:0098843) postsynaptic endocytic zone membrane(GO:0098844) |
| 0.0 | 0.2 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.0 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.0 | GO:0060187 | cell pole(GO:0060187) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.2 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 0.2 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.1 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 0.2 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.1 | GO:0008892 | guanine deaminase activity(GO:0008892) |
| 0.0 | 0.2 | GO:0042019 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 0.1 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.0 | 0.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.2 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.1 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.1 | GO:0031403 | lithium ion binding(GO:0031403) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.2 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.0 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.0 | 0.1 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.0 | GO:0030395 | lactose binding(GO:0030395) |
| 0.0 | 0.1 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.1 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.0 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 0.0 | 0.1 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.1 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 0.0 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.2 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.1 | GO:0050659 | chondroitin 4-sulfotransferase activity(GO:0047756) N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.0 | 0.0 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.1 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.0 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.0 | 0.1 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |