Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
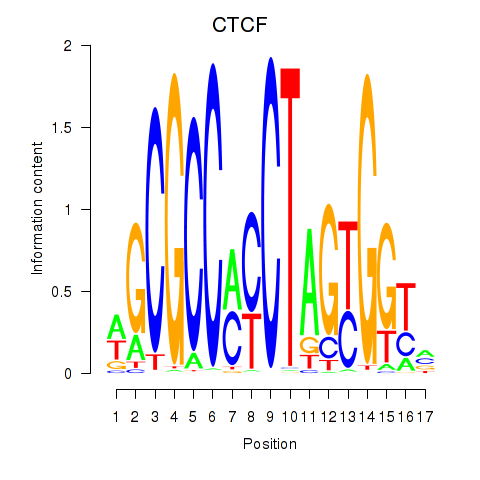
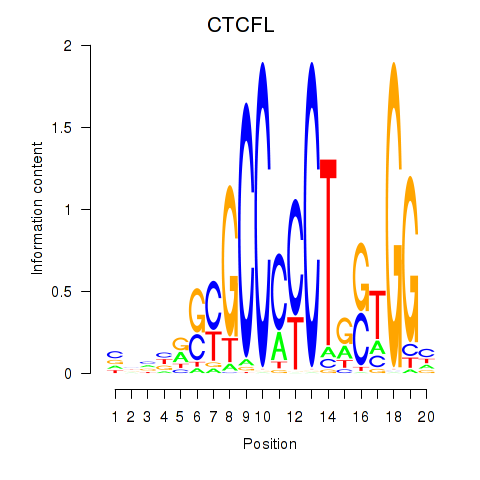
Results for CTCF_CTCFL
Z-value: 0.35
Transcription factors associated with CTCF_CTCFL
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CTCF
|
ENSG00000102974.10 | CCCTC-binding factor |
|
CTCFL
|
ENSG00000124092.8 | CCCTC-binding factor like |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CTCFL | hg19_v2_chr20_-_56100155_56100163 | -0.91 | 8.8e-02 | Click! |
| CTCF | hg19_v2_chr16_+_67596310_67596353 | -0.25 | 7.5e-01 | Click! |
Activity profile of CTCF_CTCFL motif
Sorted Z-values of CTCF_CTCFL motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_302918 | 0.68 |
ENST00000599994.1
|
AC187652.1
|
Protein LOC100996433 |
| chr15_+_45722727 | 0.66 |
ENST00000396650.2
ENST00000558435.1 ENST00000344300.3 |
C15orf48
|
chromosome 15 open reading frame 48 |
| chr18_+_43753500 | 0.39 |
ENST00000587591.1
ENST00000588730.1 |
C18orf25
|
chromosome 18 open reading frame 25 |
| chr7_+_105172612 | 0.29 |
ENST00000493041.1
|
RINT1
|
RAD50 interactor 1 |
| chr4_-_41884620 | 0.25 |
ENST00000504870.1
|
LINC00682
|
long intergenic non-protein coding RNA 682 |
| chr20_-_30311703 | 0.24 |
ENST00000450273.1
ENST00000456404.1 ENST00000420488.1 ENST00000439267.1 |
BCL2L1
|
BCL2-like 1 |
| chr16_+_29127282 | 0.24 |
ENST00000562902.1
|
RP11-426C22.5
|
RP11-426C22.5 |
| chr5_-_136834982 | 0.23 |
ENST00000510689.1
ENST00000394945.1 |
SPOCK1
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1 |
| chr4_+_7194247 | 0.23 |
ENST00000507866.2
|
SORCS2
|
sortilin-related VPS10 domain containing receptor 2 |
| chr19_-_19051927 | 0.22 |
ENST00000600077.1
|
HOMER3
|
homer homolog 3 (Drosophila) |
| chr11_+_60691924 | 0.21 |
ENST00000544065.1
ENST00000453848.2 ENST00000005286.4 |
TMEM132A
|
transmembrane protein 132A |
| chr4_+_89378261 | 0.21 |
ENST00000264350.3
|
HERC5
|
HECT and RLD domain containing E3 ubiquitin protein ligase 5 |
| chr12_+_82752275 | 0.21 |
ENST00000248306.3
|
METTL25
|
methyltransferase like 25 |
| chr9_+_100174232 | 0.20 |
ENST00000355295.4
|
TDRD7
|
tudor domain containing 7 |
| chr15_-_44487408 | 0.20 |
ENST00000402883.1
ENST00000417257.1 |
FRMD5
|
FERM domain containing 5 |
| chr9_+_100174344 | 0.19 |
ENST00000422139.2
|
TDRD7
|
tudor domain containing 7 |
| chr6_+_32821924 | 0.19 |
ENST00000374859.2
ENST00000453265.2 |
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr6_-_32821599 | 0.18 |
ENST00000354258.4
|
TAP1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr2_+_218933972 | 0.18 |
ENST00000374155.3
|
RUFY4
|
RUN and FYVE domain containing 4 |
| chr11_+_73675873 | 0.18 |
ENST00000537753.1
ENST00000542350.1 |
DNAJB13
|
DnaJ (Hsp40) homolog, subfamily B, member 13 |
| chr5_-_139726181 | 0.18 |
ENST00000507104.1
ENST00000230990.6 |
HBEGF
|
heparin-binding EGF-like growth factor |
| chr20_+_48807351 | 0.17 |
ENST00000303004.3
|
CEBPB
|
CCAAT/enhancer binding protein (C/EBP), beta |
| chr16_-_2390704 | 0.17 |
ENST00000301732.5
ENST00000382381.3 |
ABCA3
|
ATP-binding cassette, sub-family A (ABC1), member 3 |
| chrX_+_23801280 | 0.17 |
ENST00000379251.3
ENST00000379253.3 ENST00000379254.1 ENST00000379270.4 |
SAT1
|
spermidine/spermine N1-acetyltransferase 1 |
| chr22_-_50964558 | 0.17 |
ENST00000535425.1
ENST00000439934.1 |
SCO2
|
SCO2 cytochrome c oxidase assembly protein |
| chr14_+_54976603 | 0.17 |
ENST00000557317.1
|
CGRRF1
|
cell growth regulator with ring finger domain 1 |
| chr16_-_11350036 | 0.17 |
ENST00000332029.2
|
SOCS1
|
suppressor of cytokine signaling 1 |
| chr18_-_21166841 | 0.16 |
ENST00000269228.5
|
NPC1
|
Niemann-Pick disease, type C1 |
| chr17_-_61778528 | 0.15 |
ENST00000584645.1
|
LIMD2
|
LIM domain containing 2 |
| chr17_+_7591747 | 0.15 |
ENST00000534050.1
|
WRAP53
|
WD repeat containing, antisense to TP53 |
| chr2_-_154335300 | 0.15 |
ENST00000325926.3
|
RPRM
|
reprimo, TP53 dependent G2 arrest mediator candidate |
| chr10_+_104154229 | 0.15 |
ENST00000428099.1
ENST00000369966.3 |
NFKB2
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr2_+_20866424 | 0.15 |
ENST00000272224.3
|
GDF7
|
growth differentiation factor 7 |
| chr11_-_66103932 | 0.15 |
ENST00000311320.4
|
RIN1
|
Ras and Rab interactor 1 |
| chr10_-_6019552 | 0.15 |
ENST00000379977.3
ENST00000397251.3 ENST00000397248.2 |
IL15RA
|
interleukin 15 receptor, alpha |
| chr7_-_27219849 | 0.15 |
ENST00000396344.4
|
HOXA10
|
homeobox A10 |
| chr20_+_3801162 | 0.15 |
ENST00000379573.2
ENST00000379567.2 ENST00000455742.1 ENST00000246041.2 |
AP5S1
|
adaptor-related protein complex 5, sigma 1 subunit |
| chr3_-_48723268 | 0.15 |
ENST00000439518.1
ENST00000416649.2 ENST00000341520.4 ENST00000294129.2 |
NCKIPSD
|
NCK interacting protein with SH3 domain |
| chr15_+_63340775 | 0.15 |
ENST00000559281.1
ENST00000317516.7 |
TPM1
|
tropomyosin 1 (alpha) |
| chr19_-_19051993 | 0.14 |
ENST00000594794.1
ENST00000355887.6 ENST00000392351.3 ENST00000596482.1 |
HOMER3
|
homer homolog 3 (Drosophila) |
| chr3_+_48507210 | 0.14 |
ENST00000433541.1
ENST00000422277.2 ENST00000436480.2 ENST00000444177.1 |
TREX1
|
three prime repair exonuclease 1 |
| chr12_-_7245125 | 0.14 |
ENST00000542285.1
ENST00000540610.1 |
C1R
|
complement component 1, r subcomponent |
| chr17_+_7591639 | 0.14 |
ENST00000396463.2
|
WRAP53
|
WD repeat containing, antisense to TP53 |
| chr20_-_30539773 | 0.13 |
ENST00000202017.4
|
PDRG1
|
p53 and DNA-damage regulated 1 |
| chr15_+_63340734 | 0.13 |
ENST00000560959.1
|
TPM1
|
tropomyosin 1 (alpha) |
| chr19_-_3547305 | 0.13 |
ENST00000589063.1
|
MFSD12
|
major facilitator superfamily domain containing 12 |
| chr2_+_232316906 | 0.13 |
ENST00000370380.2
|
AC017104.2
|
Uncharacterized protein |
| chr4_+_56814968 | 0.13 |
ENST00000422247.2
|
CEP135
|
centrosomal protein 135kDa |
| chr11_-_44972299 | 0.13 |
ENST00000528473.1
|
TP53I11
|
tumor protein p53 inducible protein 11 |
| chr11_-_8285405 | 0.13 |
ENST00000335790.3
ENST00000534484.1 |
LMO1
|
LIM domain only 1 (rhombotin 1) |
| chr16_+_84002234 | 0.13 |
ENST00000305202.4
|
NECAB2
|
N-terminal EF-hand calcium binding protein 2 |
| chr11_-_6704513 | 0.13 |
ENST00000532203.1
ENST00000288937.6 |
MRPL17
|
mitochondrial ribosomal protein L17 |
| chr2_-_136875712 | 0.12 |
ENST00000241393.3
|
CXCR4
|
chemokine (C-X-C motif) receptor 4 |
| chr12_-_7245018 | 0.12 |
ENST00000543835.1
ENST00000535233.2 |
C1R
|
complement component 1, r subcomponent |
| chr11_+_2421718 | 0.12 |
ENST00000380996.5
ENST00000333256.6 ENST00000380992.1 ENST00000437110.1 ENST00000435795.1 |
TSSC4
|
tumor suppressing subtransferable candidate 4 |
| chr11_-_321340 | 0.12 |
ENST00000526811.1
|
IFITM3
|
interferon induced transmembrane protein 3 |
| chr22_+_22723969 | 0.12 |
ENST00000390295.2
|
IGLV7-46
|
immunoglobulin lambda variable 7-46 (gene/pseudogene) |
| chr16_-_22012419 | 0.12 |
ENST00000537222.2
ENST00000424898.2 ENST00000286143.6 |
PDZD9
|
PDZ domain containing 9 |
| chr5_+_172571445 | 0.12 |
ENST00000231668.9
ENST00000351486.5 ENST00000352523.6 ENST00000393770.4 |
BNIP1
|
BCL2/adenovirus E1B 19kDa interacting protein 1 |
| chr4_-_41216492 | 0.12 |
ENST00000503503.1
ENST00000509446.1 ENST00000503264.1 ENST00000508707.1 ENST00000508593.1 |
APBB2
|
amyloid beta (A4) precursor protein-binding, family B, member 2 |
| chr17_-_79917645 | 0.12 |
ENST00000477214.1
|
NOTUM
|
notum pectinacetylesterase homolog (Drosophila) |
| chr2_-_69064324 | 0.12 |
ENST00000415448.1
|
AC097495.3
|
AC097495.3 |
| chr12_-_56320887 | 0.12 |
ENST00000398213.4
|
WIBG
|
within bgcn homolog (Drosophila) |
| chr6_+_32811885 | 0.12 |
ENST00000458296.1
ENST00000413039.1 ENST00000429600.1 ENST00000412095.1 ENST00000415067.1 ENST00000395330.1 |
TAPSAR1
PSMB9
|
TAP1 and PSMB8 antisense RNA 1 proteasome (prosome, macropain) subunit, beta type, 9 |
| chr8_+_22022653 | 0.11 |
ENST00000354870.5
ENST00000397816.3 ENST00000306349.8 |
BMP1
|
bone morphogenetic protein 1 |
| chr11_-_66104237 | 0.11 |
ENST00000530056.1
|
RIN1
|
Ras and Rab interactor 1 |
| chr12_+_58176525 | 0.11 |
ENST00000543727.1
ENST00000540550.1 ENST00000323833.8 ENST00000350762.5 ENST00000550559.1 ENST00000548851.1 ENST00000434359.1 ENST00000457189.1 |
TSFM
|
Ts translation elongation factor, mitochondrial |
| chr20_+_49411523 | 0.11 |
ENST00000371608.2
|
BCAS4
|
breast carcinoma amplified sequence 4 |
| chr20_-_1447467 | 0.11 |
ENST00000353088.2
ENST00000350991.4 |
NSFL1C
|
NSFL1 (p97) cofactor (p47) |
| chr12_-_7245152 | 0.11 |
ENST00000542220.2
|
C1R
|
complement component 1, r subcomponent |
| chr20_+_49411543 | 0.11 |
ENST00000609336.1
ENST00000445038.1 |
BCAS4
|
breast carcinoma amplified sequence 4 |
| chr17_+_41561317 | 0.11 |
ENST00000540306.1
ENST00000262415.3 ENST00000605777.1 |
DHX8
|
DEAH (Asp-Glu-Ala-His) box polypeptide 8 |
| chr20_+_43992094 | 0.11 |
ENST00000453003.1
|
SYS1
|
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae) |
| chr7_-_105926058 | 0.11 |
ENST00000417537.1
|
NAMPT
|
nicotinamide phosphoribosyltransferase |
| chr18_-_26941900 | 0.11 |
ENST00000577674.1
|
CTD-2515C13.2
|
CTD-2515C13.2 |
| chr2_-_191878681 | 0.11 |
ENST00000409465.1
|
STAT1
|
signal transducer and activator of transcription 1, 91kDa |
| chr10_-_6019984 | 0.11 |
ENST00000525219.2
|
IL15RA
|
interleukin 15 receptor, alpha |
| chr14_+_23341513 | 0.10 |
ENST00000546834.1
|
LRP10
|
low density lipoprotein receptor-related protein 10 |
| chr19_-_48018203 | 0.10 |
ENST00000595227.1
ENST00000593761.1 ENST00000263354.3 |
NAPA
|
N-ethylmaleimide-sensitive factor attachment protein, alpha |
| chr2_+_27309605 | 0.10 |
ENST00000260599.6
ENST00000260598.5 ENST00000429697.1 |
KHK
|
ketohexokinase (fructokinase) |
| chr7_+_6629621 | 0.10 |
ENST00000344417.5
|
C7orf26
|
chromosome 7 open reading frame 26 |
| chr12_-_30907822 | 0.10 |
ENST00000540436.1
|
CAPRIN2
|
caprin family member 2 |
| chr16_+_48657361 | 0.10 |
ENST00000565072.1
|
RP11-42I10.1
|
RP11-42I10.1 |
| chr22_-_30783356 | 0.10 |
ENST00000382363.3
|
RNF215
|
ring finger protein 215 |
| chr1_+_156890418 | 0.10 |
ENST00000337428.7
|
LRRC71
|
leucine rich repeat containing 71 |
| chr18_-_48346415 | 0.10 |
ENST00000431965.2
ENST00000436348.2 |
MRO
|
maestro |
| chr11_+_44117219 | 0.10 |
ENST00000532479.1
ENST00000527014.1 |
EXT2
|
exostosin glycosyltransferase 2 |
| chr17_-_34122596 | 0.10 |
ENST00000250144.8
|
MMP28
|
matrix metallopeptidase 28 |
| chr12_-_54785074 | 0.10 |
ENST00000338010.5
ENST00000550774.1 |
ZNF385A
|
zinc finger protein 385A |
| chr19_-_39832563 | 0.10 |
ENST00000599274.1
|
CTC-246B18.10
|
CTC-246B18.10 |
| chr2_+_220306238 | 0.10 |
ENST00000435853.1
|
SPEG
|
SPEG complex locus |
| chr2_-_191878874 | 0.10 |
ENST00000392322.3
ENST00000392323.2 ENST00000424722.1 ENST00000361099.3 |
STAT1
|
signal transducer and activator of transcription 1, 91kDa |
| chr19_+_42363917 | 0.10 |
ENST00000598742.1
|
RPS19
|
ribosomal protein S19 |
| chr12_-_7245080 | 0.10 |
ENST00000541042.1
ENST00000540242.1 |
C1R
|
complement component 1, r subcomponent |
| chr20_+_3767547 | 0.10 |
ENST00000344256.6
ENST00000379598.5 |
CDC25B
|
cell division cycle 25B |
| chr3_-_48598547 | 0.09 |
ENST00000536104.1
ENST00000452531.1 |
PFKFB4
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 |
| chr19_+_12949251 | 0.09 |
ENST00000251472.4
|
MAST1
|
microtubule associated serine/threonine kinase 1 |
| chr11_-_57335280 | 0.09 |
ENST00000287156.4
|
UBE2L6
|
ubiquitin-conjugating enzyme E2L 6 |
| chr19_+_41313017 | 0.09 |
ENST00000595621.1
ENST00000595051.1 |
EGLN2
|
egl-9 family hypoxia-inducible factor 2 |
| chr2_-_232395169 | 0.09 |
ENST00000305141.4
|
NMUR1
|
neuromedin U receptor 1 |
| chr14_+_23025534 | 0.09 |
ENST00000557595.1
|
AE000662.92
|
Uncharacterized protein |
| chr12_-_54785054 | 0.09 |
ENST00000352268.6
ENST00000549962.1 |
ZNF385A
|
zinc finger protein 385A |
| chr8_-_30670053 | 0.09 |
ENST00000518564.1
|
PPP2CB
|
protein phosphatase 2, catalytic subunit, beta isozyme |
| chr1_-_36615051 | 0.09 |
ENST00000373163.1
|
TRAPPC3
|
trafficking protein particle complex 3 |
| chr10_+_13141441 | 0.09 |
ENST00000263036.5
|
OPTN
|
optineurin |
| chr6_+_138188378 | 0.09 |
ENST00000420009.1
|
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr9_-_139258159 | 0.09 |
ENST00000371739.3
|
DNLZ
|
DNL-type zinc finger |
| chr9_+_131464767 | 0.09 |
ENST00000291906.4
|
PKN3
|
protein kinase N3 |
| chr17_-_7518145 | 0.09 |
ENST00000250113.7
ENST00000571597.1 |
FXR2
|
fragile X mental retardation, autosomal homolog 2 |
| chr16_+_69345243 | 0.09 |
ENST00000254950.11
|
VPS4A
|
vacuolar protein sorting 4 homolog A (S. cerevisiae) |
| chr12_+_50505762 | 0.09 |
ENST00000550487.1
ENST00000317943.2 |
COX14
|
cytochrome c oxidase assembly homolog 14 (S. cerevisiae) |
| chr10_+_13141585 | 0.09 |
ENST00000378764.2
|
OPTN
|
optineurin |
| chr20_+_49411431 | 0.09 |
ENST00000358791.5
ENST00000262591.5 |
BCAS4
|
breast carcinoma amplified sequence 4 |
| chr1_-_35325318 | 0.09 |
ENST00000423898.1
ENST00000456842.1 |
SMIM12
|
small integral membrane protein 12 |
| chr12_+_122356488 | 0.09 |
ENST00000397454.2
|
WDR66
|
WD repeat domain 66 |
| chr19_+_18699599 | 0.09 |
ENST00000450195.2
|
C19orf60
|
chromosome 19 open reading frame 60 |
| chr11_+_118826999 | 0.09 |
ENST00000264031.2
|
UPK2
|
uroplakin 2 |
| chr17_+_74864476 | 0.09 |
ENST00000301618.4
ENST00000569840.2 |
MGAT5B
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase, isozyme B |
| chr6_-_44225231 | 0.08 |
ENST00000538577.1
ENST00000537814.1 ENST00000393810.1 ENST00000393812.3 |
SLC35B2
|
solute carrier family 35 (adenosine 3'-phospho 5'-phosphosulfate transporter), member B2 |
| chr12_+_13197218 | 0.08 |
ENST00000197268.8
|
KIAA1467
|
KIAA1467 |
| chr16_+_67360712 | 0.08 |
ENST00000569499.2
ENST00000329956.6 ENST00000561948.1 |
LRRC36
|
leucine rich repeat containing 36 |
| chr17_-_1090599 | 0.08 |
ENST00000544583.2
|
ABR
|
active BCR-related |
| chr20_-_30310797 | 0.08 |
ENST00000422920.1
|
BCL2L1
|
BCL2-like 1 |
| chr22_-_50964849 | 0.08 |
ENST00000543927.1
ENST00000423348.1 |
SCO2
|
SCO2 cytochrome c oxidase assembly protein |
| chr14_+_24630465 | 0.08 |
ENST00000557894.1
ENST00000559284.1 ENST00000560275.1 |
IRF9
|
interferon regulatory factor 9 |
| chr10_+_12391685 | 0.08 |
ENST00000378845.1
|
CAMK1D
|
calcium/calmodulin-dependent protein kinase ID |
| chr7_+_129007964 | 0.08 |
ENST00000460109.1
ENST00000474594.1 ENST00000446212.1 |
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr6_+_32811861 | 0.08 |
ENST00000453426.1
|
TAPSAR1
|
TAP1 and PSMB8 antisense RNA 1 |
| chr5_+_150827143 | 0.08 |
ENST00000243389.3
ENST00000517945.1 ENST00000521925.1 |
SLC36A1
|
solute carrier family 36 (proton/amino acid symporter), member 1 |
| chr12_+_110338323 | 0.08 |
ENST00000312777.5
ENST00000536408.2 |
TCHP
|
trichoplein, keratin filament binding |
| chr3_-_126373929 | 0.08 |
ENST00000523403.1
ENST00000524230.2 |
TXNRD3
|
thioredoxin reductase 3 |
| chr12_+_110338063 | 0.08 |
ENST00000405876.4
|
TCHP
|
trichoplein, keratin filament binding |
| chr16_+_48657337 | 0.08 |
ENST00000568150.1
ENST00000564212.1 |
RP11-42I10.1
|
RP11-42I10.1 |
| chr7_+_6144514 | 0.08 |
ENST00000306177.5
ENST00000465073.2 |
USP42
|
ubiquitin specific peptidase 42 |
| chr16_+_22308717 | 0.08 |
ENST00000299853.5
ENST00000564209.1 ENST00000565358.1 ENST00000418581.2 ENST00000564883.1 ENST00000359210.4 ENST00000563024.1 |
POLR3E
|
polymerase (RNA) III (DNA directed) polypeptide E (80kD) |
| chr6_+_138188551 | 0.08 |
ENST00000237289.4
ENST00000433680.1 |
TNFAIP3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr22_-_30783075 | 0.08 |
ENST00000215798.6
|
RNF215
|
ring finger protein 215 |
| chr17_+_65027509 | 0.08 |
ENST00000375684.1
|
AC005544.1
|
Uncharacterized protein |
| chr11_+_64781575 | 0.08 |
ENST00000246747.4
ENST00000529384.1 |
ARL2
|
ADP-ribosylation factor-like 2 |
| chr20_-_1447547 | 0.08 |
ENST00000476071.1
|
NSFL1C
|
NSFL1 (p97) cofactor (p47) |
| chr17_-_35716019 | 0.08 |
ENST00000591148.1
ENST00000394406.2 ENST00000394403.1 |
ACACA
|
acetyl-CoA carboxylase alpha |
| chr22_-_50963976 | 0.08 |
ENST00000252785.3
ENST00000395693.3 |
SCO2
|
SCO2 cytochrome c oxidase assembly protein |
| chr10_+_81107216 | 0.08 |
ENST00000394579.3
ENST00000225174.3 |
PPIF
|
peptidylprolyl isomerase F |
| chr19_-_19739321 | 0.08 |
ENST00000588461.1
|
LPAR2
|
lysophosphatidic acid receptor 2 |
| chr19_-_1848451 | 0.08 |
ENST00000170168.4
|
REXO1
|
REX1, RNA exonuclease 1 homolog (S. cerevisiae) |
| chr11_-_117103208 | 0.08 |
ENST00000320934.3
ENST00000530269.1 |
PCSK7
|
proprotein convertase subtilisin/kexin type 7 |
| chr12_+_50505963 | 0.08 |
ENST00000550654.1
ENST00000548985.1 |
COX14
|
cytochrome c oxidase assembly homolog 14 (S. cerevisiae) |
| chr12_+_48357340 | 0.08 |
ENST00000256686.6
ENST00000549288.1 ENST00000552561.1 ENST00000546749.1 ENST00000552546.1 ENST00000550552.1 |
TMEM106C
|
transmembrane protein 106C |
| chr1_-_145589424 | 0.08 |
ENST00000334513.5
|
NUDT17
|
nudix (nucleoside diphosphate linked moiety X)-type motif 17 |
| chr3_+_156393349 | 0.07 |
ENST00000473702.1
|
TIPARP
|
TCDD-inducible poly(ADP-ribose) polymerase |
| chr17_-_48277552 | 0.07 |
ENST00000507689.1
|
COL1A1
|
collagen, type I, alpha 1 |
| chr16_+_67679069 | 0.07 |
ENST00000545661.1
|
RLTPR
|
RGD motif, leucine rich repeats, tropomodulin domain and proline-rich containing |
| chr19_-_55652290 | 0.07 |
ENST00000589745.1
|
TNNT1
|
troponin T type 1 (skeletal, slow) |
| chr19_-_50990785 | 0.07 |
ENST00000595005.1
|
CTD-2545M3.8
|
CTD-2545M3.8 |
| chr1_-_91317072 | 0.07 |
ENST00000435649.2
ENST00000443802.1 |
RP4-665J23.1
|
RP4-665J23.1 |
| chr2_-_219157219 | 0.07 |
ENST00000445635.1
ENST00000413976.1 |
TMBIM1
|
transmembrane BAX inhibitor motif containing 1 |
| chr10_+_112257596 | 0.07 |
ENST00000369583.3
|
DUSP5
|
dual specificity phosphatase 5 |
| chr5_-_141257954 | 0.07 |
ENST00000456271.1
ENST00000394536.3 ENST00000503492.1 ENST00000287008.3 |
PCDH1
|
protocadherin 1 |
| chr3_-_194354010 | 0.07 |
ENST00000381975.3
ENST00000347147.4 ENST00000473092.1 ENST00000392432.2 ENST00000273580.7 |
TMEM44
|
transmembrane protein 44 |
| chr9_+_34646651 | 0.07 |
ENST00000378842.3
|
GALT
|
galactose-1-phosphate uridylyltransferase |
| chr11_-_65149422 | 0.07 |
ENST00000526432.1
ENST00000527174.1 |
SLC25A45
|
solute carrier family 25, member 45 |
| chr17_+_19912640 | 0.07 |
ENST00000395527.4
ENST00000583482.2 ENST00000583528.1 ENST00000583463.1 |
SPECC1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr7_-_45957011 | 0.07 |
ENST00000417621.1
|
IGFBP3
|
insulin-like growth factor binding protein 3 |
| chr9_+_34646624 | 0.07 |
ENST00000450095.2
ENST00000556278.1 |
GALT
GALT
|
galactose-1-phosphate uridylyltransferase Uncharacterized protein |
| chr19_+_16222439 | 0.07 |
ENST00000300935.3
|
RAB8A
|
RAB8A, member RAS oncogene family |
| chr16_+_31724618 | 0.07 |
ENST00000530881.1
ENST00000529515.1 |
ZNF720
|
zinc finger protein 720 |
| chr15_+_63340647 | 0.07 |
ENST00000404484.4
|
TPM1
|
tropomyosin 1 (alpha) |
| chr12_+_106751436 | 0.07 |
ENST00000228347.4
|
POLR3B
|
polymerase (RNA) III (DNA directed) polypeptide B |
| chr5_-_137475071 | 0.07 |
ENST00000265191.2
|
NME5
|
NME/NM23 family member 5 |
| chr9_+_100263912 | 0.07 |
ENST00000259365.4
|
TMOD1
|
tropomodulin 1 |
| chr19_-_36001386 | 0.07 |
ENST00000461300.1
|
DMKN
|
dermokine |
| chrX_+_118108571 | 0.07 |
ENST00000304778.7
|
LONRF3
|
LON peptidase N-terminal domain and ring finger 3 |
| chr7_-_45956856 | 0.07 |
ENST00000428530.1
|
IGFBP3
|
insulin-like growth factor binding protein 3 |
| chr2_-_196933536 | 0.07 |
ENST00000312428.6
ENST00000410072.1 |
DNAH7
|
dynein, axonemal, heavy chain 7 |
| chr2_+_233497931 | 0.07 |
ENST00000264059.3
|
EFHD1
|
EF-hand domain family, member D1 |
| chr15_+_75940218 | 0.07 |
ENST00000308527.5
|
SNX33
|
sorting nexin 33 |
| chr11_-_68611721 | 0.07 |
ENST00000561996.1
|
CPT1A
|
carnitine palmitoyltransferase 1A (liver) |
| chr16_+_67678818 | 0.07 |
ENST00000334583.6
|
RLTPR
|
RGD motif, leucine rich repeats, tropomodulin domain and proline-rich containing |
| chr10_-_79789291 | 0.07 |
ENST00000372371.3
|
POLR3A
|
polymerase (RNA) III (DNA directed) polypeptide A, 155kDa |
| chr17_+_79071365 | 0.07 |
ENST00000576756.1
|
BAIAP2
|
BAI1-associated protein 2 |
| chr11_-_65359947 | 0.07 |
ENST00000597463.1
|
AP001362.1
|
Uncharacterized protein |
| chr5_+_140180635 | 0.07 |
ENST00000522353.2
ENST00000532566.2 |
PCDHA3
|
protocadherin alpha 3 |
| chr10_-_103603677 | 0.07 |
ENST00000358038.3
|
KCNIP2
|
Kv channel interacting protein 2 |
| chr12_-_120703523 | 0.07 |
ENST00000267257.7
ENST00000228307.7 ENST00000424649.2 |
PXN
|
paxillin |
| chr10_-_15762124 | 0.07 |
ENST00000378076.3
|
ITGA8
|
integrin, alpha 8 |
| chr9_+_140317802 | 0.07 |
ENST00000341349.2
ENST00000392815.2 |
NOXA1
|
NADPH oxidase activator 1 |
| chr20_-_36793663 | 0.07 |
ENST00000536701.1
ENST00000536724.1 |
TGM2
|
transglutaminase 2 |
| chr1_+_193091080 | 0.07 |
ENST00000367435.3
|
CDC73
|
cell division cycle 73 |
| chr3_-_197476560 | 0.07 |
ENST00000273582.5
|
KIAA0226
|
KIAA0226 |
| chr8_+_22022223 | 0.07 |
ENST00000306385.5
|
BMP1
|
bone morphogenetic protein 1 |
| chr19_-_13213954 | 0.07 |
ENST00000590974.1
|
LYL1
|
lymphoblastic leukemia derived sequence 1 |
| chr16_+_14927538 | 0.07 |
ENST00000287667.7
|
NOMO1
|
NODAL modulator 1 |
| chr15_+_63340553 | 0.06 |
ENST00000334895.5
|
TPM1
|
tropomyosin 1 (alpha) |
| chr19_+_4791722 | 0.06 |
ENST00000269856.3
|
FEM1A
|
fem-1 homolog a (C. elegans) |
| chr14_-_77608121 | 0.06 |
ENST00000319374.4
|
ZDHHC22
|
zinc finger, DHHC-type containing 22 |
| chr10_+_6244829 | 0.06 |
ENST00000317350.4
ENST00000379785.1 ENST00000379782.3 ENST00000360521.2 ENST00000379775.4 |
PFKFB3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr15_+_66679155 | 0.06 |
ENST00000307102.5
|
MAP2K1
|
mitogen-activated protein kinase kinase 1 |
| chr19_+_5690207 | 0.06 |
ENST00000347512.3
|
RPL36
|
ribosomal protein L36 |
| chrX_-_27417088 | 0.06 |
ENST00000608735.1
ENST00000422048.1 |
RP11-268G12.1
|
RP11-268G12.1 |
| chr11_+_58938903 | 0.06 |
ENST00000532982.1
|
DTX4
|
deltex homolog 4 (Drosophila) |
Network of associatons between targets according to the STRING database.
First level regulatory network of CTCF_CTCFL
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.1 | 0.2 | GO:0072573 | regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) tolerance induction to lipopolysaccharide(GO:0072573) negative regulation of CD40 signaling pathway(GO:2000349) |
| 0.1 | 0.3 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.1 | 0.2 | GO:0032917 | polyamine acetylation(GO:0032917) spermidine acetylation(GO:0032918) |
| 0.1 | 0.4 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.1 | 0.2 | GO:0072308 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 0.0 | 0.3 | GO:0072425 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.0 | 0.1 | GO:0006258 | UDP-glucose catabolic process(GO:0006258) |
| 0.0 | 0.2 | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 0.0 | 0.3 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.0 | 0.1 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.1 | GO:0060168 | positive regulation of adenosine receptor signaling pathway(GO:0060168) |
| 0.0 | 0.3 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.1 | GO:0048210 | Golgi vesicle fusion to target membrane(GO:0048210) |
| 0.0 | 0.1 | GO:1990697 | protein depalmitoleylation(GO:1990697) |
| 0.0 | 0.2 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.0 | 0.2 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.2 | GO:1902162 | regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.1 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 | 0.1 | GO:0046963 | 3'-phosphoadenosine 5'-phosphosulfate transport(GO:0046963) 3'-phospho-5'-adenylyl sulfate transmembrane transport(GO:1902559) |
| 0.0 | 0.3 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0018153 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.0 | 0.2 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.2 | GO:0035992 | tendon cell differentiation(GO:0035990) tendon formation(GO:0035992) |
| 0.0 | 0.1 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.0 | 0.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.4 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.1 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.0 | 0.1 | GO:0044026 | DNA hypermethylation(GO:0044026) |
| 0.0 | 0.1 | GO:0034402 | recruitment of 3'-end processing factors to RNA polymerase II holoenzyme complex(GO:0034402) |
| 0.0 | 0.2 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 0.1 | GO:0010725 | regulation of primitive erythrocyte differentiation(GO:0010725) eosinophil fate commitment(GO:0035854) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.2 | GO:0010533 | regulation of activation of Janus kinase activity(GO:0010533) |
| 0.0 | 0.1 | GO:0038098 | sequestering of BMP from receptor via BMP binding(GO:0038098) |
| 0.0 | 0.1 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.0 | 0.1 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.1 | GO:1902045 | negative regulation of Fas signaling pathway(GO:1902045) positive regulation of blood vessel remodeling(GO:2000504) |
| 0.0 | 0.5 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.2 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.1 | GO:0044800 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 | 0.0 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.0 | GO:2000276 | negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.0 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.1 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.0 | 0.1 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.0 | 0.0 | GO:0021644 | vagus nerve morphogenesis(GO:0021644) |
| 0.0 | 0.1 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.0 | 0.0 | GO:0009213 | pyrimidine nucleoside triphosphate catabolic process(GO:0009149) pyrimidine deoxyribonucleoside triphosphate catabolic process(GO:0009213) |
| 0.0 | 0.1 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.1 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.1 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.4 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.0 | GO:0060532 | bronchus cartilage development(GO:0060532) lung smooth muscle development(GO:0061145) |
| 0.0 | 0.2 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.2 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.1 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.0 | 0.0 | GO:0090579 | transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.0 | 0.1 | GO:0003420 | regulation of growth plate cartilage chondrocyte proliferation(GO:0003420) |
| 0.0 | 0.0 | GO:0016333 | morphogenesis of follicular epithelium(GO:0016333) establishment or maintenance of polarity of follicular epithelium(GO:0016334) establishment of planar polarity of follicular epithelium(GO:0042247) |
| 0.0 | 0.2 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 | 0.0 | GO:2000724 | nuclear fragmentation involved in apoptotic nuclear change(GO:0030264) positive regulation of cardiac vascular smooth muscle cell differentiation(GO:2000724) |
| 0.0 | 0.1 | GO:0045819 | positive regulation of glycogen catabolic process(GO:0045819) |
| 0.0 | 0.1 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.0 | 0.1 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.0 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.1 | GO:0006311 | meiotic gene conversion(GO:0006311) male meiosis chromosome segregation(GO:0007060) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0097233 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.0 | 0.3 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.2 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.0 | 0.3 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.2 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.0 | 0.2 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.0 | 0.1 | GO:0001534 | radial spoke(GO:0001534) |
| 0.0 | 0.3 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.4 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.4 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.1 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.1 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.1 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.0 | 0.1 | GO:0034678 | integrin alpha8-beta1 complex(GO:0034678) |
| 0.0 | 0.1 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 0.2 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) polyamine binding(GO:0019808) |
| 0.0 | 0.1 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.0 | 0.1 | GO:1990699 | palmitoleyl hydrolase activity(GO:1990699) |
| 0.0 | 0.1 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 0.0 | 0.3 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.1 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.3 | GO:0015440 | peptide-transporting ATPase activity(GO:0015440) TAP binding(GO:0046977) TAP1 binding(GO:0046978) |
| 0.0 | 0.3 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.0 | 0.1 | GO:0004513 | neolactotetraosylceramide alpha-2,3-sialyltransferase activity(GO:0004513) lactosylceramide alpha-2,3-sialyltransferase activity(GO:0047291) |
| 0.0 | 0.1 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.0 | 0.1 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 0.3 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.2 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.1 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.0 | 0.1 | GO:0031862 | prostanoid receptor binding(GO:0031862) |
| 0.0 | 0.1 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.1 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.1 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.0 | 0.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.0 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.1 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.1 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.0 | 0.3 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.0 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.1 | GO:0035651 | AP-1 adaptor complex binding(GO:0035650) AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.3 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.0 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.0 | 0.0 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.0 | 0.0 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.0 | 0.3 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.1 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.4 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.3 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |