Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
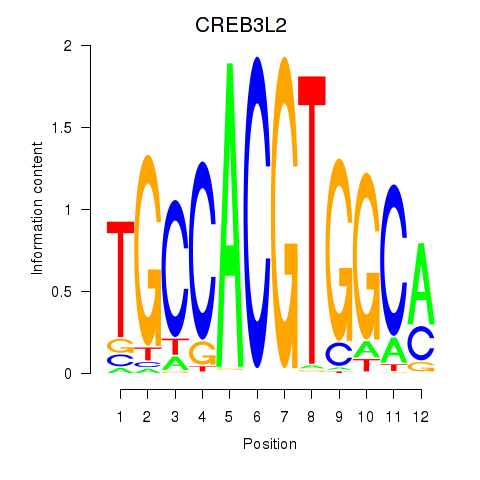
Results for CREB3L2
Z-value: 0.54
Transcription factors associated with CREB3L2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CREB3L2
|
ENSG00000182158.10 | cAMP responsive element binding protein 3 like 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CREB3L2 | hg19_v2_chr7_-_137686791_137686821 | 0.74 | 2.6e-01 | Click! |
Activity profile of CREB3L2 motif
Sorted Z-values of CREB3L2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_96928516 | 0.60 |
ENST00000602703.1
|
RP11-2B6.3
|
RP11-2B6.3 |
| chr2_-_27341966 | 0.46 |
ENST00000402394.1
ENST00000402550.1 ENST00000260595.5 |
CGREF1
|
cell growth regulator with EF-hand domain 1 |
| chr14_+_74551650 | 0.40 |
ENST00000554938.1
|
LIN52
|
lin-52 homolog (C. elegans) |
| chr5_+_71475449 | 0.40 |
ENST00000504492.1
|
MAP1B
|
microtubule-associated protein 1B |
| chr20_+_18118703 | 0.39 |
ENST00000464792.1
|
CSRP2BP
|
CSRP2 binding protein |
| chr2_-_27341765 | 0.37 |
ENST00000405600.1
|
CGREF1
|
cell growth regulator with EF-hand domain 1 |
| chr7_+_16793160 | 0.29 |
ENST00000262067.4
|
TSPAN13
|
tetraspanin 13 |
| chr8_-_99129338 | 0.28 |
ENST00000520507.1
|
HRSP12
|
heat-responsive protein 12 |
| chr7_-_8302298 | 0.27 |
ENST00000446305.1
|
ICA1
|
islet cell autoantigen 1, 69kDa |
| chr5_+_92228 | 0.26 |
ENST00000512035.1
|
CTD-2231H16.1
|
CTD-2231H16.1 |
| chr4_+_119200215 | 0.26 |
ENST00000602573.1
|
SNHG8
|
small nucleolar RNA host gene 8 (non-protein coding) |
| chr14_-_100070363 | 0.26 |
ENST00000380243.4
|
CCDC85C
|
coiled-coil domain containing 85C |
| chr9_+_92219919 | 0.25 |
ENST00000252506.6
ENST00000375769.1 |
GADD45G
|
growth arrest and DNA-damage-inducible, gamma |
| chr3_-_123603137 | 0.24 |
ENST00000360304.3
ENST00000359169.1 ENST00000346322.5 ENST00000360772.3 |
MYLK
|
myosin light chain kinase |
| chr12_+_121416437 | 0.23 |
ENST00000402929.1
ENST00000535955.1 ENST00000538626.1 ENST00000543427.1 |
HNF1A
|
HNF1 homeobox A |
| chr8_-_144679264 | 0.23 |
ENST00000531953.1
ENST00000526133.1 |
EEF1D
|
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chr9_+_37079888 | 0.21 |
ENST00000429493.1
ENST00000593237.1 ENST00000588557.1 ENST00000430809.1 ENST00000592157.1 |
RP11-220I1.1
|
RP11-220I1.1 |
| chr9_+_131902283 | 0.21 |
ENST00000436883.1
ENST00000414510.1 |
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr2_-_198364581 | 0.20 |
ENST00000428204.1
|
HSPD1
|
heat shock 60kDa protein 1 (chaperonin) |
| chr2_-_27341872 | 0.20 |
ENST00000312734.4
|
CGREF1
|
cell growth regulator with EF-hand domain 1 |
| chr3_-_155572164 | 0.20 |
ENST00000392845.3
ENST00000359479.3 |
SLC33A1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr8_-_99129384 | 0.20 |
ENST00000521560.1
ENST00000254878.3 |
HRSP12
|
heat-responsive protein 12 |
| chr5_-_55008072 | 0.20 |
ENST00000512208.1
|
SLC38A9
|
solute carrier family 38, member 9 |
| chr1_+_29213584 | 0.19 |
ENST00000343067.4
ENST00000356093.2 ENST00000398863.2 ENST00000373800.3 ENST00000349460.4 |
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) |
| chr6_-_136871957 | 0.19 |
ENST00000354570.3
|
MAP7
|
microtubule-associated protein 7 |
| chr10_+_89419370 | 0.19 |
ENST00000361175.4
ENST00000456849.1 |
PAPSS2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr6_+_127588020 | 0.19 |
ENST00000309649.3
ENST00000610162.1 ENST00000610153.1 ENST00000608991.1 ENST00000480444.1 |
RNF146
|
ring finger protein 146 |
| chr5_+_41904431 | 0.19 |
ENST00000381647.2
|
C5orf51
|
chromosome 5 open reading frame 51 |
| chr13_+_35516390 | 0.18 |
ENST00000540320.1
ENST00000400445.3 ENST00000310336.4 |
NBEA
|
neurobeachin |
| chr19_+_49458107 | 0.18 |
ENST00000539787.1
ENST00000345358.7 ENST00000391871.3 ENST00000415969.2 ENST00000354470.3 ENST00000506183.1 ENST00000293288.8 |
BAX
|
BCL2-associated X protein |
| chr15_+_75315896 | 0.17 |
ENST00000342932.3
ENST00000564923.1 ENST00000569562.1 ENST00000568649.1 |
PPCDC
|
phosphopantothenoylcysteine decarboxylase |
| chr18_+_56530794 | 0.17 |
ENST00000590285.1
ENST00000586085.1 ENST00000589288.1 |
ZNF532
|
zinc finger protein 532 |
| chr18_+_56531584 | 0.17 |
ENST00000590287.1
|
ZNF532
|
zinc finger protein 532 |
| chr4_+_119199904 | 0.17 |
ENST00000602483.1
ENST00000602819.1 |
SNHG8
|
small nucleolar RNA host gene 8 (non-protein coding) |
| chr16_-_21314360 | 0.17 |
ENST00000219599.3
ENST00000576703.1 |
CRYM
|
crystallin, mu |
| chr19_+_36705504 | 0.17 |
ENST00000456324.1
|
ZNF146
|
zinc finger protein 146 |
| chr22_-_36903069 | 0.16 |
ENST00000216187.6
ENST00000423980.1 |
FOXRED2
|
FAD-dependent oxidoreductase domain containing 2 |
| chr17_+_39968926 | 0.16 |
ENST00000585664.1
ENST00000585922.1 ENST00000429461.1 |
FKBP10
|
FK506 binding protein 10, 65 kDa |
| chr5_+_43603229 | 0.16 |
ENST00000344920.4
ENST00000512996.2 |
NNT
|
nicotinamide nucleotide transhydrogenase |
| chr8_+_22102611 | 0.16 |
ENST00000306433.4
|
POLR3D
|
polymerase (RNA) III (DNA directed) polypeptide D, 44kDa |
| chr10_+_131265443 | 0.16 |
ENST00000306010.7
|
MGMT
|
O-6-methylguanine-DNA methyltransferase |
| chrX_+_131157322 | 0.16 |
ENST00000481105.1
ENST00000354719.6 ENST00000394335.2 |
MST4
|
Serine/threonine-protein kinase MST4 |
| chr10_+_70715884 | 0.16 |
ENST00000354185.4
|
DDX21
|
DEAD (Asp-Glu-Ala-Asp) box helicase 21 |
| chr6_-_27840099 | 0.16 |
ENST00000328488.2
|
HIST1H3I
|
histone cluster 1, H3i |
| chr19_+_36706024 | 0.15 |
ENST00000443387.2
|
ZNF146
|
zinc finger protein 146 |
| chr7_+_100464760 | 0.15 |
ENST00000200457.4
|
TRIP6
|
thyroid hormone receptor interactor 6 |
| chr2_+_172544294 | 0.15 |
ENST00000358002.6
ENST00000435234.1 ENST00000443458.1 ENST00000412370.1 |
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chr16_+_57220193 | 0.15 |
ENST00000564435.1
ENST00000562959.1 ENST00000394420.4 ENST00000568505.2 ENST00000537866.1 |
RSPRY1
|
ring finger and SPRY domain containing 1 |
| chr4_+_119199864 | 0.15 |
ENST00000602414.1
ENST00000602520.1 |
SNHG8
|
small nucleolar RNA host gene 8 (non-protein coding) |
| chr14_-_74551172 | 0.14 |
ENST00000553458.1
|
ALDH6A1
|
aldehyde dehydrogenase 6 family, member A1 |
| chr3_+_12525931 | 0.14 |
ENST00000446004.1
ENST00000314571.7 ENST00000454502.2 ENST00000383797.5 ENST00000402228.3 ENST00000284995.6 ENST00000444864.1 |
TSEN2
|
TSEN2 tRNA splicing endonuclease subunit |
| chr17_-_30185946 | 0.14 |
ENST00000579741.1
|
COPRS
|
coordinator of PRMT5, differentiation stimulator |
| chr20_-_61051026 | 0.14 |
ENST00000252997.2
|
GATA5
|
GATA binding protein 5 |
| chr14_+_50065459 | 0.14 |
ENST00000318317.4
|
LRR1
|
leucine rich repeat protein 1 |
| chr2_-_69614373 | 0.14 |
ENST00000361060.5
ENST00000357308.4 |
GFPT1
|
glutamine--fructose-6-phosphate transaminase 1 |
| chr19_+_1249869 | 0.14 |
ENST00000591446.2
|
MIDN
|
midnolin |
| chr22_-_24093267 | 0.14 |
ENST00000341976.3
|
ZNF70
|
zinc finger protein 70 |
| chr20_-_62168672 | 0.14 |
ENST00000217185.2
|
PTK6
|
protein tyrosine kinase 6 |
| chr15_+_76135622 | 0.14 |
ENST00000338677.4
ENST00000267938.4 ENST00000569423.1 |
UBE2Q2
|
ubiquitin-conjugating enzyme E2Q family member 2 |
| chr3_+_45636219 | 0.13 |
ENST00000273317.4
|
LIMD1
|
LIM domains containing 1 |
| chr14_+_96342729 | 0.13 |
ENST00000504119.1
|
LINC00617
|
long intergenic non-protein coding RNA 617 |
| chr2_-_47572105 | 0.13 |
ENST00000419035.1
ENST00000448713.1 ENST00000450550.1 ENST00000413185.2 |
AC073283.4
|
AC073283.4 |
| chr19_-_47290535 | 0.13 |
ENST00000412532.2
|
SLC1A5
|
solute carrier family 1 (neutral amino acid transporter), member 5 |
| chr19_-_42759300 | 0.13 |
ENST00000222329.4
|
ERF
|
Ets2 repressor factor |
| chr6_-_144329531 | 0.13 |
ENST00000429150.1
ENST00000392309.1 ENST00000416623.1 ENST00000392307.1 |
PLAGL1
|
pleiomorphic adenoma gene-like 1 |
| chr11_-_57282349 | 0.13 |
ENST00000528450.1
|
SLC43A1
|
solute carrier family 43 (amino acid system L transporter), member 1 |
| chr12_-_56123444 | 0.13 |
ENST00000546457.1
ENST00000549117.1 |
CD63
|
CD63 molecule |
| chr8_+_22102626 | 0.13 |
ENST00000519237.1
ENST00000397802.4 |
POLR3D
|
polymerase (RNA) III (DNA directed) polypeptide D, 44kDa |
| chr14_-_74551096 | 0.13 |
ENST00000350259.4
|
ALDH6A1
|
aldehyde dehydrogenase 6 family, member A1 |
| chr17_+_74723031 | 0.13 |
ENST00000586200.1
|
METTL23
|
methyltransferase like 23 |
| chr16_+_333152 | 0.12 |
ENST00000219406.6
ENST00000404312.1 ENST00000456379.1 |
PDIA2
|
protein disulfide isomerase family A, member 2 |
| chr1_+_46016465 | 0.12 |
ENST00000434299.1
|
AKR1A1
|
aldo-keto reductase family 1, member A1 (aldehyde reductase) |
| chr20_+_18118486 | 0.12 |
ENST00000432901.3
|
PET117
|
PET117 homolog (S. cerevisiae) |
| chr10_+_6392278 | 0.12 |
ENST00000391437.1
|
DKFZP667F0711
|
DKFZP667F0711 |
| chr19_-_36705763 | 0.12 |
ENST00000591473.1
|
ZNF565
|
zinc finger protein 565 |
| chrX_+_48433326 | 0.12 |
ENST00000376755.1
|
RBM3
|
RNA binding motif (RNP1, RRM) protein 3 |
| chr17_-_37309480 | 0.12 |
ENST00000539608.1
|
PLXDC1
|
plexin domain containing 1 |
| chr8_+_22225041 | 0.12 |
ENST00000289952.5
ENST00000524285.1 |
SLC39A14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr7_-_6523688 | 0.12 |
ENST00000490996.1
|
KDELR2
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2 |
| chr15_-_93353028 | 0.12 |
ENST00000557398.2
|
FAM174B
|
family with sequence similarity 174, member B |
| chr6_-_117923520 | 0.12 |
ENST00000368498.2
|
GOPC
|
golgi-associated PDZ and coiled-coil motif containing |
| chr1_+_100435535 | 0.11 |
ENST00000427993.2
|
SLC35A3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3 |
| chr16_+_85061367 | 0.11 |
ENST00000538274.1
ENST00000258180.3 |
KIAA0513
|
KIAA0513 |
| chr3_+_158519654 | 0.11 |
ENST00000415822.2
ENST00000392813.4 ENST00000264266.8 |
MFSD1
|
major facilitator superfamily domain containing 1 |
| chr5_+_133984462 | 0.11 |
ENST00000398844.2
ENST00000322887.4 |
SEC24A
|
SEC24 family member A |
| chr17_-_17109579 | 0.11 |
ENST00000321560.3
|
PLD6
|
phospholipase D family, member 6 |
| chr12_+_510795 | 0.11 |
ENST00000412006.2
|
CCDC77
|
coiled-coil domain containing 77 |
| chr2_+_46926326 | 0.11 |
ENST00000394861.2
|
SOCS5
|
suppressor of cytokine signaling 5 |
| chr20_-_62168714 | 0.11 |
ENST00000542869.1
|
PTK6
|
protein tyrosine kinase 6 |
| chr12_-_49318715 | 0.11 |
ENST00000444214.2
|
FKBP11
|
FK506 binding protein 11, 19 kDa |
| chr14_-_61190754 | 0.10 |
ENST00000216513.4
|
SIX4
|
SIX homeobox 4 |
| chr22_+_38864041 | 0.10 |
ENST00000216014.4
ENST00000409006.3 |
KDELR3
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 |
| chr2_+_172544011 | 0.10 |
ENST00000508530.1
|
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chr22_-_36903101 | 0.10 |
ENST00000397224.4
|
FOXRED2
|
FAD-dependent oxidoreductase domain containing 2 |
| chr8_+_22224760 | 0.10 |
ENST00000359741.5
ENST00000520644.1 ENST00000240095.6 |
SLC39A14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr12_+_510742 | 0.10 |
ENST00000239830.4
|
CCDC77
|
coiled-coil domain containing 77 |
| chr11_+_67776012 | 0.10 |
ENST00000539229.1
|
ALDH3B1
|
aldehyde dehydrogenase 3 family, member B1 |
| chr1_+_101361782 | 0.10 |
ENST00000357650.4
|
SLC30A7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr12_+_46777450 | 0.09 |
ENST00000551503.1
|
RP11-96H19.1
|
RP11-96H19.1 |
| chr5_-_138725594 | 0.09 |
ENST00000302125.8
|
MZB1
|
marginal zone B and B1 cell-specific protein |
| chr12_-_76953573 | 0.09 |
ENST00000549646.1
ENST00000550628.1 ENST00000553139.1 ENST00000261183.3 ENST00000393250.4 |
OSBPL8
|
oxysterol binding protein-like 8 |
| chr10_-_120840309 | 0.09 |
ENST00000369144.3
|
EIF3A
|
eukaryotic translation initiation factor 3, subunit A |
| chr17_+_65821780 | 0.09 |
ENST00000321892.4
ENST00000335221.5 ENST00000306378.6 |
BPTF
|
bromodomain PHD finger transcription factor |
| chr9_+_131901710 | 0.09 |
ENST00000524946.2
|
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr17_+_39969183 | 0.09 |
ENST00000321562.4
|
FKBP10
|
FK506 binding protein 10, 65 kDa |
| chr12_+_121416340 | 0.09 |
ENST00000257555.6
ENST00000400024.2 |
HNF1A
|
HNF1 homeobox A |
| chr15_-_72565340 | 0.09 |
ENST00000568360.1
|
PARP6
|
poly (ADP-ribose) polymerase family, member 6 |
| chr12_-_104532062 | 0.09 |
ENST00000240055.3
|
NFYB
|
nuclear transcription factor Y, beta |
| chr6_+_127587755 | 0.09 |
ENST00000368314.1
ENST00000476956.1 ENST00000609447.1 ENST00000356799.2 ENST00000477776.1 ENST00000609944.1 |
RNF146
|
ring finger protein 146 |
| chr5_+_149737202 | 0.09 |
ENST00000451292.1
ENST00000377797.3 ENST00000445265.2 ENST00000323668.7 ENST00000439160.2 ENST00000394269.3 ENST00000427724.2 ENST00000504761.2 ENST00000513346.1 ENST00000515516.1 |
TCOF1
|
Treacher Collins-Franceschetti syndrome 1 |
| chr2_+_74425689 | 0.09 |
ENST00000394053.2
ENST00000409804.1 ENST00000264090.4 ENST00000394050.3 ENST00000409601.1 |
MTHFD2
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase |
| chr1_-_154193009 | 0.09 |
ENST00000368518.1
ENST00000368519.1 ENST00000368521.5 |
C1orf43
|
chromosome 1 open reading frame 43 |
| chr12_+_121416489 | 0.09 |
ENST00000541395.1
ENST00000544413.1 |
HNF1A
|
HNF1 homeobox A |
| chr16_+_66914264 | 0.08 |
ENST00000311765.2
ENST00000568869.1 ENST00000561704.1 ENST00000568398.1 ENST00000566776.1 |
PDP2
|
pyruvate dehyrogenase phosphatase catalytic subunit 2 |
| chr22_-_36902522 | 0.08 |
ENST00000397223.4
|
FOXRED2
|
FAD-dependent oxidoreductase domain containing 2 |
| chr8_-_134584092 | 0.08 |
ENST00000522652.1
|
ST3GAL1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chrX_+_131157290 | 0.08 |
ENST00000394334.2
|
MST4
|
Serine/threonine-protein kinase MST4 |
| chr1_-_154193091 | 0.08 |
ENST00000362076.4
ENST00000350592.3 ENST00000368516.1 |
C1orf43
|
chromosome 1 open reading frame 43 |
| chr6_-_117923610 | 0.08 |
ENST00000535237.1
ENST00000052569.6 |
GOPC
|
golgi-associated PDZ and coiled-coil motif containing |
| chr16_+_2533020 | 0.08 |
ENST00000562105.1
|
TBC1D24
|
TBC1 domain family, member 24 |
| chr8_-_144679296 | 0.08 |
ENST00000317198.6
|
EEF1D
|
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chr2_+_172543967 | 0.08 |
ENST00000534253.2
ENST00000263811.4 ENST00000397119.3 ENST00000410079.3 ENST00000438879.1 |
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chr1_+_29213678 | 0.08 |
ENST00000347529.3
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) |
| chr6_+_122720681 | 0.08 |
ENST00000368455.4
ENST00000452194.1 |
HSF2
|
heat shock transcription factor 2 |
| chr1_+_249200395 | 0.08 |
ENST00000355360.4
ENST00000329291.5 ENST00000539153.1 |
PGBD2
|
piggyBac transposable element derived 2 |
| chr1_+_110162448 | 0.08 |
ENST00000342115.4
ENST00000469039.2 ENST00000474459.1 ENST00000528667.1 |
AMPD2
|
adenosine monophosphate deaminase 2 |
| chr5_-_1799932 | 0.08 |
ENST00000382647.7
ENST00000505059.2 |
MRPL36
|
mitochondrial ribosomal protein L36 |
| chr5_-_143550159 | 0.08 |
ENST00000448443.2
ENST00000513112.1 ENST00000519064.1 ENST00000274496.5 |
YIPF5
|
Yip1 domain family, member 5 |
| chr2_+_169312725 | 0.08 |
ENST00000392687.4
|
CERS6
|
ceramide synthase 6 |
| chr12_+_50355647 | 0.07 |
ENST00000293599.6
|
AQP5
|
aquaporin 5 |
| chr19_+_10947251 | 0.07 |
ENST00000592854.1
|
C19orf38
|
chromosome 19 open reading frame 38 |
| chr20_+_57267669 | 0.07 |
ENST00000356091.6
|
NPEPL1
|
aminopeptidase-like 1 |
| chr1_-_202896310 | 0.07 |
ENST00000367261.3
|
KLHL12
|
kelch-like family member 12 |
| chr17_+_55163075 | 0.07 |
ENST00000571629.1
ENST00000570423.1 ENST00000575186.1 ENST00000573085.1 ENST00000572814.1 |
AKAP1
|
A kinase (PRKA) anchor protein 1 |
| chr9_+_131901661 | 0.07 |
ENST00000423100.1
|
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr9_-_133769225 | 0.07 |
ENST00000343079.1
|
QRFP
|
pyroglutamylated RFamide peptide |
| chr4_+_79697495 | 0.07 |
ENST00000502871.1
ENST00000335016.5 |
BMP2K
|
BMP2 inducible kinase |
| chr8_+_9009296 | 0.07 |
ENST00000521718.1
|
RP11-10A14.4
|
Uncharacterized protein |
| chr18_+_56806701 | 0.07 |
ENST00000587834.1
|
SEC11C
|
SEC11 homolog C (S. cerevisiae) |
| chr17_+_19552036 | 0.07 |
ENST00000581518.1
ENST00000395575.2 ENST00000584332.2 ENST00000339618.4 ENST00000579855.1 |
ALDH3A2
|
aldehyde dehydrogenase 3 family, member A2 |
| chr5_+_68710906 | 0.06 |
ENST00000325631.5
ENST00000454295.2 |
MARVELD2
|
MARVEL domain containing 2 |
| chr6_-_36953833 | 0.06 |
ENST00000538808.1
ENST00000460219.1 ENST00000373616.5 ENST00000373627.5 |
MTCH1
|
mitochondrial carrier 1 |
| chr12_-_56122761 | 0.06 |
ENST00000552164.1
ENST00000420846.3 ENST00000257857.4 |
CD63
|
CD63 molecule |
| chr15_+_62359175 | 0.06 |
ENST00000355522.5
|
C2CD4A
|
C2 calcium-dependent domain containing 4A |
| chr17_+_48450575 | 0.06 |
ENST00000338165.4
ENST00000393271.2 ENST00000511519.2 |
EME1
|
essential meiotic structure-specific endonuclease 1 |
| chr16_+_70613770 | 0.06 |
ENST00000429149.2
ENST00000563721.2 |
IL34
|
interleukin 34 |
| chr2_-_211036051 | 0.06 |
ENST00000418791.1
ENST00000452086.1 ENST00000281772.9 |
KANSL1L
|
KAT8 regulatory NSL complex subunit 1-like |
| chr1_+_32479430 | 0.06 |
ENST00000327300.7
ENST00000492989.1 |
KHDRBS1
|
KH domain containing, RNA binding, signal transduction associated 1 |
| chr9_-_136242956 | 0.06 |
ENST00000371989.3
ENST00000485435.2 |
SURF4
|
surfeit 4 |
| chr4_+_37455536 | 0.06 |
ENST00000381980.4
ENST00000508175.1 |
C4orf19
|
chromosome 4 open reading frame 19 |
| chr1_-_114447412 | 0.06 |
ENST00000369567.1
ENST00000369566.3 |
AP4B1
|
adaptor-related protein complex 4, beta 1 subunit |
| chr22_-_43411106 | 0.06 |
ENST00000453643.1
ENST00000263246.3 ENST00000337959.4 |
PACSIN2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr2_+_169312350 | 0.06 |
ENST00000305747.6
|
CERS6
|
ceramide synthase 6 |
| chr1_-_231376867 | 0.06 |
ENST00000366649.2
ENST00000318906.2 ENST00000366651.3 |
C1orf131
|
chromosome 1 open reading frame 131 |
| chr17_+_74722912 | 0.06 |
ENST00000589977.1
ENST00000591571.1 ENST00000592849.1 ENST00000586738.1 ENST00000588783.1 ENST00000588563.1 ENST00000586752.1 ENST00000588302.1 ENST00000590964.1 ENST00000341249.6 ENST00000588822.1 |
METTL23
|
methyltransferase like 23 |
| chr19_-_47164386 | 0.06 |
ENST00000391916.2
ENST00000410105.2 |
DACT3
|
dishevelled-binding antagonist of beta-catenin 3 |
| chr3_+_51428704 | 0.06 |
ENST00000323686.4
|
RBM15B
|
RNA binding motif protein 15B |
| chr1_-_145039949 | 0.06 |
ENST00000313382.9
|
PDE4DIP
|
phosphodiesterase 4D interacting protein |
| chr11_+_111957497 | 0.05 |
ENST00000375549.3
ENST00000528182.1 ENST00000528048.1 ENST00000528021.1 ENST00000526592.1 ENST00000525291.1 |
SDHD
|
succinate dehydrogenase complex, subunit D, integral membrane protein |
| chr1_+_46016703 | 0.05 |
ENST00000481885.1
ENST00000351829.4 ENST00000471651.1 |
AKR1A1
|
aldo-keto reductase family 1, member A1 (aldehyde reductase) |
| chr11_-_36310958 | 0.05 |
ENST00000532705.1
ENST00000263401.5 ENST00000452374.2 |
COMMD9
|
COMM domain containing 9 |
| chr3_+_148709310 | 0.05 |
ENST00000484197.1
ENST00000492285.2 ENST00000461191.1 |
GYG1
|
glycogenin 1 |
| chr3_+_169490606 | 0.05 |
ENST00000349841.5
|
MYNN
|
myoneurin |
| chr7_-_1595871 | 0.05 |
ENST00000319010.5
|
TMEM184A
|
transmembrane protein 184A |
| chr12_+_57624085 | 0.05 |
ENST00000553474.1
|
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr19_+_34663551 | 0.05 |
ENST00000586157.1
|
LSM14A
|
LSM14A, SCD6 homolog A (S. cerevisiae) |
| chr22_-_43253189 | 0.05 |
ENST00000437119.2
ENST00000429508.2 ENST00000454099.1 ENST00000263245.5 |
ARFGAP3
|
ADP-ribosylation factor GTPase activating protein 3 |
| chr2_+_172543919 | 0.05 |
ENST00000452242.1
ENST00000340296.4 |
DYNC1I2
|
dynein, cytoplasmic 1, intermediate chain 2 |
| chr8_-_143696833 | 0.05 |
ENST00000356613.2
|
ARC
|
activity-regulated cytoskeleton-associated protein |
| chr20_+_2633269 | 0.05 |
ENST00000445139.1
|
NOP56
|
NOP56 ribonucleoprotein |
| chr6_+_3259148 | 0.05 |
ENST00000419065.2
ENST00000473000.2 ENST00000451246.2 ENST00000454610.2 |
PSMG4
|
proteasome (prosome, macropain) assembly chaperone 4 |
| chr17_+_75452447 | 0.05 |
ENST00000591472.2
|
SEPT9
|
septin 9 |
| chr11_+_45944190 | 0.05 |
ENST00000401752.1
ENST00000389968.3 ENST00000325468.5 ENST00000536139.1 |
GYLTL1B
|
glycosyltransferase-like 1B |
| chr21_-_40720995 | 0.05 |
ENST00000380749.5
|
HMGN1
|
high mobility group nucleosome binding domain 1 |
| chr10_-_71930222 | 0.05 |
ENST00000458634.2
ENST00000373239.2 ENST00000373242.2 ENST00000373241.4 |
SAR1A
|
SAR1 homolog A (S. cerevisiae) |
| chrX_+_48432892 | 0.05 |
ENST00000376759.3
ENST00000430348.2 |
RBM3
|
RNA binding motif (RNP1, RRM) protein 3 |
| chr8_+_99129513 | 0.05 |
ENST00000522319.1
ENST00000401707.2 |
POP1
|
processing of precursor 1, ribonuclease P/MRP subunit (S. cerevisiae) |
| chr1_-_113498616 | 0.04 |
ENST00000433570.4
ENST00000538576.1 ENST00000458229.1 |
SLC16A1
|
solute carrier family 16 (monocarboxylate transporter), member 1 |
| chr20_+_61584026 | 0.04 |
ENST00000370351.4
ENST00000370349.3 |
SLC17A9
|
solute carrier family 17 (vesicular nucleotide transporter), member 9 |
| chr2_-_197036289 | 0.04 |
ENST00000263955.4
|
STK17B
|
serine/threonine kinase 17b |
| chr11_-_777467 | 0.04 |
ENST00000397472.2
ENST00000524550.1 ENST00000319863.8 ENST00000526325.1 ENST00000442059.2 |
PDDC1
|
Parkinson disease 7 domain containing 1 |
| chr3_-_57583185 | 0.04 |
ENST00000463880.1
|
ARF4
|
ADP-ribosylation factor 4 |
| chr17_-_7137857 | 0.04 |
ENST00000005340.5
|
DVL2
|
dishevelled segment polarity protein 2 |
| chr5_+_68711023 | 0.04 |
ENST00000515844.1
|
MARVELD2
|
MARVEL domain containing 2 |
| chr16_+_699319 | 0.04 |
ENST00000549091.1
ENST00000293879.4 |
WDR90
|
WD repeat domain 90 |
| chr19_+_10812108 | 0.04 |
ENST00000250237.5
ENST00000592254.1 |
QTRT1
|
queuine tRNA-ribosyltransferase 1 |
| chr19_-_2721336 | 0.04 |
ENST00000588128.1
|
DIRAS1
|
DIRAS family, GTP-binding RAS-like 1 |
| chr2_-_73340146 | 0.04 |
ENST00000258098.6
|
RAB11FIP5
|
RAB11 family interacting protein 5 (class I) |
| chr11_+_1430629 | 0.04 |
ENST00000528596.1
|
BRSK2
|
BR serine/threonine kinase 2 |
| chr16_+_88923494 | 0.04 |
ENST00000567895.1
ENST00000301021.3 ENST00000565504.1 ENST00000567312.1 ENST00000568583.1 ENST00000561840.1 |
TRAPPC2L
|
trafficking protein particle complex 2-like |
| chr5_-_1799965 | 0.04 |
ENST00000508987.1
|
MRPL36
|
mitochondrial ribosomal protein L36 |
| chr2_+_201676908 | 0.04 |
ENST00000409226.1
ENST00000452790.2 |
BZW1
|
basic leucine zipper and W2 domains 1 |
| chr15_-_90645679 | 0.04 |
ENST00000539790.1
ENST00000559482.1 ENST00000330062.3 |
IDH2
|
isocitrate dehydrogenase 2 (NADP+), mitochondrial |
| chr1_+_116915855 | 0.04 |
ENST00000295598.5
|
ATP1A1
|
ATPase, Na+/K+ transporting, alpha 1 polypeptide |
| chr6_-_144329384 | 0.04 |
ENST00000417959.2
|
PLAGL1
|
pleiomorphic adenoma gene-like 1 |
| chr12_+_57623477 | 0.04 |
ENST00000557487.1
ENST00000555634.1 ENST00000556689.1 |
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr17_+_57408994 | 0.04 |
ENST00000312655.4
|
YPEL2
|
yippee-like 2 (Drosophila) |
| chr15_-_98417780 | 0.04 |
ENST00000503874.3
|
LINC00923
|
long intergenic non-protein coding RNA 923 |
| chr5_-_143550241 | 0.04 |
ENST00000522203.1
|
YIPF5
|
Yip1 domain family, member 5 |
| chr4_+_80584903 | 0.04 |
ENST00000506460.1
|
RP11-452C8.1
|
RP11-452C8.1 |
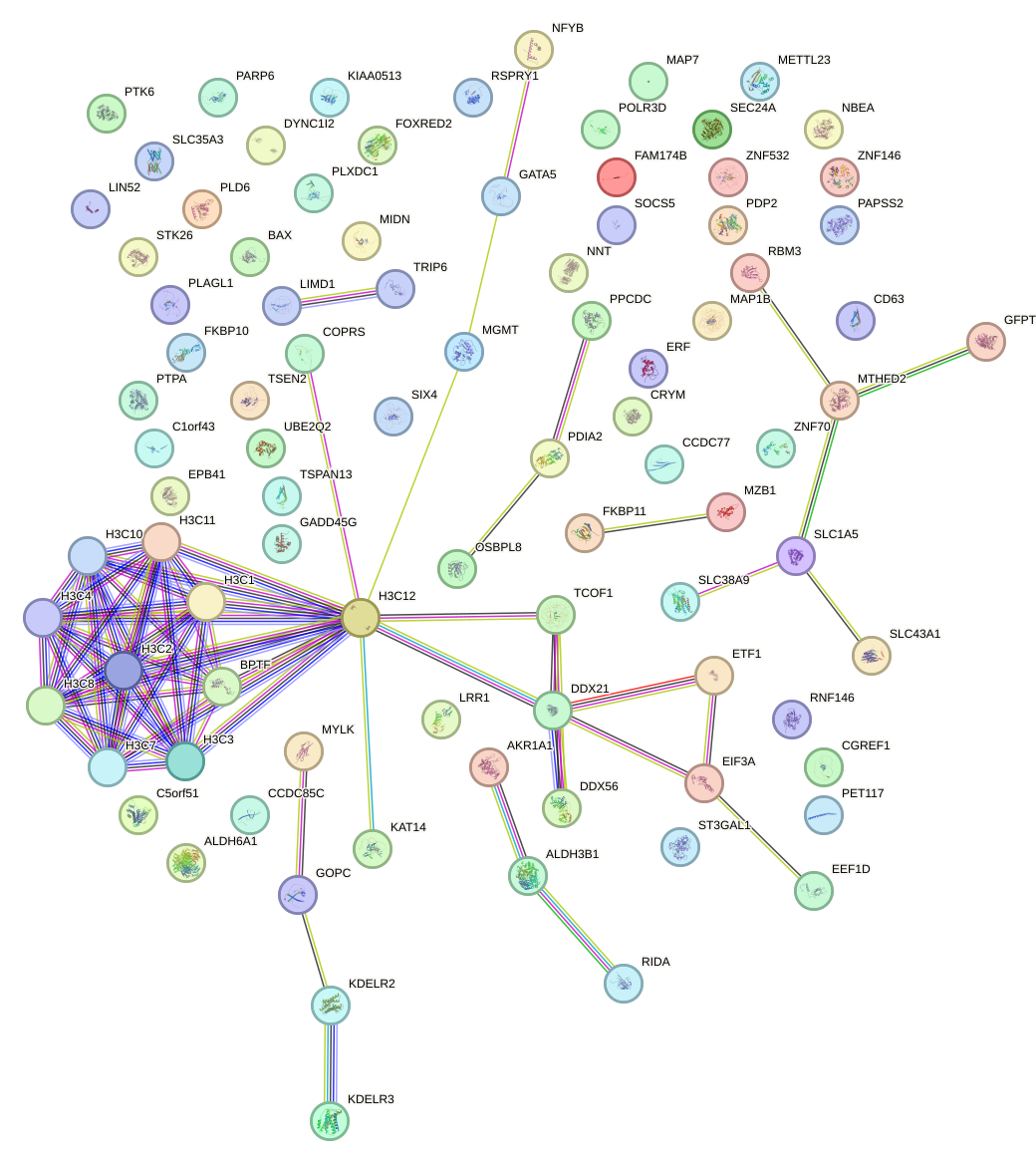
Network of associatons between targets according to the STRING database.
First level regulatory network of CREB3L2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0035623 | renal glucose absorption(GO:0035623) |
| 0.1 | 0.3 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.1 | 0.2 | GO:0045041 | B cell cytokine production(GO:0002368) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.1 | 0.2 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) Golgi to plasma membrane CFTR protein transport(GO:0043000) |
| 0.1 | 0.3 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.1 | 0.2 | GO:1902512 | B cell selection(GO:0002339) B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) positive regulation of apoptotic DNA fragmentation(GO:1902512) |
| 0.0 | 0.2 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.2 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 0.0 | 0.0 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.4 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.1 | GO:0010585 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.0 | 0.2 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.2 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.4 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.1 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.0 | 0.2 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.1 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.0 | 0.4 | GO:0061162 | establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.0 | 0.1 | GO:0046833 | positive regulation of RNA export from nucleus(GO:0046833) |
| 0.0 | 0.2 | GO:0035826 | rubidium ion transport(GO:0035826) regulation of rubidium ion transport(GO:2000680) |
| 0.0 | 0.1 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 0.2 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.1 | GO:0002378 | immunoglobulin biosynthetic process(GO:0002378) negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 0.0 | 0.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.4 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.1 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 | 0.2 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.2 | GO:1903874 | ferrous iron transport(GO:0015684) ferrous iron transmembrane transport(GO:1903874) |
| 0.0 | 0.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.0 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.0 | 0.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.0 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.3 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.1 | GO:0043614 | multi-eIF complex(GO:0043614) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.2 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.3 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.1 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.2 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.1 | 0.2 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.2 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 0.2 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.2 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.1 | GO:0005046 | KDEL sequence binding(GO:0005046) |
| 0.0 | 0.1 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.1 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.0 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.2 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.1 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.2 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.1 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.4 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.1 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 0.0 | 0.0 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.0 | 0.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.1 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.3 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.4 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |