Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
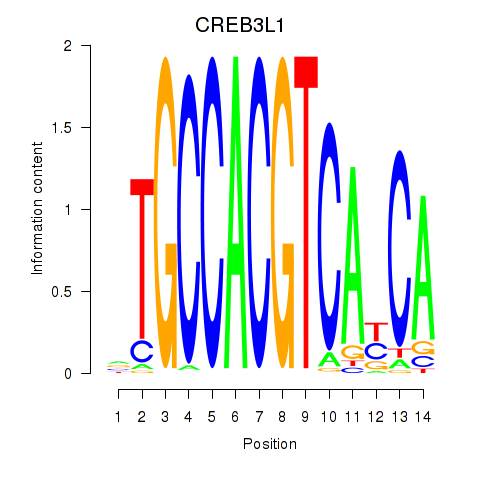
Results for CREB3L1_CREB3
Z-value: 0.82
Transcription factors associated with CREB3L1_CREB3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CREB3L1
|
ENSG00000157613.6 | cAMP responsive element binding protein 3 like 1 |
|
CREB3
|
ENSG00000107175.6 | cAMP responsive element binding protein 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CREB3 | hg19_v2_chr9_+_35732312_35732332 | -0.95 | 5.0e-02 | Click! |
| CREB3L1 | hg19_v2_chr11_+_46316677_46316719 | -0.63 | 3.7e-01 | Click! |
Activity profile of CREB3L1_CREB3 motif
Sorted Z-values of CREB3L1_CREB3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_6522922 | 1.78 |
ENST00000601673.1
|
FLJ20306
|
CDNA FLJ20306 fis, clone HEP06881; Putative uncharacterized protein FLJ20306; Uncharacterized protein |
| chr19_+_50031547 | 1.09 |
ENST00000597801.1
|
RCN3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr6_+_31916733 | 0.51 |
ENST00000483004.1
|
CFB
|
complement factor B |
| chr13_+_36920569 | 0.44 |
ENST00000379848.2
|
SPG20OS
|
SPG20 opposite strand |
| chr10_-_22292675 | 0.36 |
ENST00000376946.1
|
DNAJC1
|
DnaJ (Hsp40) homolog, subfamily C, member 1 |
| chr16_+_56642041 | 0.34 |
ENST00000245185.5
|
MT2A
|
metallothionein 2A |
| chr16_+_56642489 | 0.34 |
ENST00000561491.1
|
MT2A
|
metallothionein 2A |
| chr22_+_38864041 | 0.33 |
ENST00000216014.4
ENST00000409006.3 |
KDELR3
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 |
| chr11_+_1891380 | 0.32 |
ENST00000429923.1
ENST00000418975.1 ENST00000406638.2 |
LSP1
|
lymphocyte-specific protein 1 |
| chr12_+_113376249 | 0.32 |
ENST00000551007.1
ENST00000548514.1 |
OAS3
|
2'-5'-oligoadenylate synthetase 3, 100kDa |
| chr10_+_64893039 | 0.30 |
ENST00000277746.6
ENST00000435510.2 |
NRBF2
|
nuclear receptor binding factor 2 |
| chr7_+_72848092 | 0.29 |
ENST00000344575.3
|
FZD9
|
frizzled family receptor 9 |
| chr6_-_90062543 | 0.29 |
ENST00000435041.2
|
UBE2J1
|
ubiquitin-conjugating enzyme E2, J1 |
| chr4_+_75311019 | 0.28 |
ENST00000502307.1
|
AREG
|
amphiregulin |
| chr13_-_52027134 | 0.28 |
ENST00000311234.4
ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6
|
integrator complex subunit 6 |
| chr15_-_80695917 | 0.28 |
ENST00000559008.1
|
RP11-210M15.2
|
Uncharacterized protein |
| chr4_+_56262115 | 0.27 |
ENST00000506198.1
ENST00000381334.5 ENST00000542052.1 |
TMEM165
|
transmembrane protein 165 |
| chr9_+_126777676 | 0.26 |
ENST00000488674.2
|
LHX2
|
LIM homeobox 2 |
| chr17_+_57642886 | 0.26 |
ENST00000251241.4
ENST00000451169.2 ENST00000425628.3 ENST00000584385.1 ENST00000580030.1 |
DHX40
|
DEAH (Asp-Glu-Ala-His) box polypeptide 40 |
| chr12_+_72233487 | 0.25 |
ENST00000482439.2
ENST00000550746.1 ENST00000491063.1 ENST00000319106.8 ENST00000485960.2 ENST00000393309.3 |
TBC1D15
|
TBC1 domain family, member 15 |
| chr3_-_119182453 | 0.25 |
ENST00000491685.1
ENST00000461654.1 |
TMEM39A
|
transmembrane protein 39A |
| chr4_+_75310851 | 0.25 |
ENST00000395748.3
ENST00000264487.2 |
AREG
|
amphiregulin |
| chr1_+_145576007 | 0.24 |
ENST00000369298.1
|
PIAS3
|
protein inhibitor of activated STAT, 3 |
| chr8_-_64080945 | 0.24 |
ENST00000603538.1
|
YTHDF3-AS1
|
YTHDF3 antisense RNA 1 (head to head) |
| chr17_-_6554877 | 0.24 |
ENST00000225728.3
ENST00000575197.1 |
MED31
|
mediator complex subunit 31 |
| chr14_+_78227105 | 0.24 |
ENST00000439131.2
ENST00000355883.3 ENST00000557011.1 ENST00000556047.1 |
C14orf178
|
chromosome 14 open reading frame 178 |
| chr5_-_131132658 | 0.24 |
ENST00000514667.1
ENST00000511848.1 ENST00000510461.1 |
CTC-432M15.3
FNIP1
|
Folliculin-interacting protein 1 folliculin interacting protein 1 |
| chr11_+_12108410 | 0.24 |
ENST00000527997.1
|
RP13-631K18.5
|
RP13-631K18.5 |
| chr20_+_31595406 | 0.24 |
ENST00000170150.3
|
BPIFB2
|
BPI fold containing family B, member 2 |
| chr12_+_112451222 | 0.23 |
ENST00000552052.1
|
ERP29
|
endoplasmic reticulum protein 29 |
| chr9_+_112542572 | 0.23 |
ENST00000374530.3
|
PALM2-AKAP2
|
PALM2-AKAP2 readthrough |
| chr1_+_145575980 | 0.23 |
ENST00000393045.2
|
PIAS3
|
protein inhibitor of activated STAT, 3 |
| chr20_+_33146510 | 0.23 |
ENST00000397709.1
|
MAP1LC3A
|
microtubule-associated protein 1 light chain 3 alpha |
| chr5_+_122110691 | 0.22 |
ENST00000379516.2
ENST00000505934.1 ENST00000514949.1 |
SNX2
|
sorting nexin 2 |
| chr10_-_27444143 | 0.22 |
ENST00000477432.1
|
YME1L1
|
YME1-like 1 ATPase |
| chr12_-_93835665 | 0.21 |
ENST00000552442.1
ENST00000550657.1 |
UBE2N
|
ubiquitin-conjugating enzyme E2N |
| chr11_-_119234876 | 0.21 |
ENST00000525735.1
|
USP2
|
ubiquitin specific peptidase 2 |
| chr12_-_48276710 | 0.21 |
ENST00000550314.1
|
VDR
|
vitamin D (1,25- dihydroxyvitamin D3) receptor |
| chr5_-_131132614 | 0.21 |
ENST00000307968.7
ENST00000307954.8 |
FNIP1
|
folliculin interacting protein 1 |
| chr21_-_38445297 | 0.20 |
ENST00000430792.1
ENST00000399103.1 |
PIGP
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr3_-_139108463 | 0.20 |
ENST00000512242.1
|
COPB2
|
coatomer protein complex, subunit beta 2 (beta prime) |
| chr22_+_20850171 | 0.20 |
ENST00000445987.1
|
MED15
|
mediator complex subunit 15 |
| chr5_+_94890840 | 0.20 |
ENST00000504763.1
|
ARSK
|
arylsulfatase family, member K |
| chr2_-_202645612 | 0.20 |
ENST00000409632.2
ENST00000410052.1 ENST00000467448.1 |
ALS2
|
amyotrophic lateral sclerosis 2 (juvenile) |
| chr3_-_25824872 | 0.20 |
ENST00000308710.5
|
NGLY1
|
N-glycanase 1 |
| chr14_+_101293687 | 0.19 |
ENST00000455286.1
|
MEG3
|
maternally expressed 3 (non-protein coding) |
| chr22_+_50981079 | 0.19 |
ENST00000609268.1
|
CTA-384D8.34
|
CTA-384D8.34 |
| chr5_-_176827631 | 0.19 |
ENST00000358571.2
|
PFN3
|
profilin 3 |
| chr14_-_35344093 | 0.18 |
ENST00000382422.2
|
BAZ1A
|
bromodomain adjacent to zinc finger domain, 1A |
| chr10_-_14920599 | 0.18 |
ENST00000609399.1
|
RP11-398C13.6
|
RP11-398C13.6 |
| chr17_-_6554747 | 0.18 |
ENST00000574128.1
|
MED31
|
mediator complex subunit 31 |
| chr4_+_108746282 | 0.18 |
ENST00000503862.1
|
SGMS2
|
sphingomyelin synthase 2 |
| chr19_-_46916805 | 0.18 |
ENST00000307522.3
|
CCDC8
|
coiled-coil domain containing 8 |
| chr4_+_141445333 | 0.17 |
ENST00000507667.1
|
ELMOD2
|
ELMO/CED-12 domain containing 2 |
| chr11_+_69455855 | 0.17 |
ENST00000227507.2
ENST00000536559.1 |
CCND1
|
cyclin D1 |
| chr19_-_52097613 | 0.17 |
ENST00000301439.3
|
AC018755.1
|
HCG2008157; Uncharacterized protein; cDNA FLJ30403 fis, clone BRACE2008480 |
| chr11_-_59383617 | 0.16 |
ENST00000263847.1
|
OSBP
|
oxysterol binding protein |
| chr21_+_45138941 | 0.16 |
ENST00000398081.1
ENST00000468090.1 ENST00000291565.4 |
PDXK
|
pyridoxal (pyridoxine, vitamin B6) kinase |
| chr1_-_113498943 | 0.16 |
ENST00000369626.3
|
SLC16A1
|
solute carrier family 16 (monocarboxylate transporter), member 1 |
| chr18_-_46987000 | 0.16 |
ENST00000442713.2
ENST00000269445.6 |
DYM
|
dymeclin |
| chr18_-_47018869 | 0.16 |
ENST00000583036.1
ENST00000580261.1 |
RPL17
|
ribosomal protein L17 |
| chr4_+_6202448 | 0.16 |
ENST00000508601.1
|
RP11-586D19.1
|
RP11-586D19.1 |
| chrX_-_19689106 | 0.16 |
ENST00000379716.1
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr10_+_27444268 | 0.15 |
ENST00000375940.4
ENST00000342386.6 |
MASTL
|
microtubule associated serine/threonine kinase-like |
| chr1_+_42921761 | 0.15 |
ENST00000372562.1
|
PPCS
|
phosphopantothenoylcysteine synthetase |
| chr7_+_39663061 | 0.15 |
ENST00000005257.2
|
RALA
|
v-ral simian leukemia viral oncogene homolog A (ras related) |
| chr3_-_150264272 | 0.15 |
ENST00000491660.1
ENST00000487153.1 ENST00000239944.2 |
SERP1
|
stress-associated endoplasmic reticulum protein 1 |
| chr11_-_64646086 | 0.15 |
ENST00000320631.3
|
EHD1
|
EH-domain containing 1 |
| chr20_+_42574317 | 0.15 |
ENST00000358131.5
|
TOX2
|
TOX high mobility group box family member 2 |
| chr21_-_38445011 | 0.15 |
ENST00000464265.1
ENST00000399102.1 |
PIGP
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr21_+_44313375 | 0.14 |
ENST00000354250.2
ENST00000340344.4 |
NDUFV3
|
NADH dehydrogenase (ubiquinone) flavoprotein 3, 10kDa |
| chrX_+_153059608 | 0.14 |
ENST00000370087.1
|
SSR4
|
signal sequence receptor, delta |
| chr3_-_119182506 | 0.14 |
ENST00000468676.1
|
TMEM39A
|
transmembrane protein 39A |
| chr1_+_160313062 | 0.14 |
ENST00000294785.5
ENST00000368063.1 ENST00000437169.1 |
NCSTN
|
nicastrin |
| chr19_-_18391708 | 0.14 |
ENST00000600972.1
|
JUND
|
jun D proto-oncogene |
| chr16_+_57220049 | 0.14 |
ENST00000562439.1
|
RSPRY1
|
ring finger and SPRY domain containing 1 |
| chr15_-_52030293 | 0.14 |
ENST00000560491.1
ENST00000267838.3 |
LYSMD2
|
LysM, putative peptidoglycan-binding, domain containing 2 |
| chr19_+_50180409 | 0.14 |
ENST00000391851.4
|
PRMT1
|
protein arginine methyltransferase 1 |
| chr5_-_176730733 | 0.13 |
ENST00000504395.1
|
RAB24
|
RAB24, member RAS oncogene family |
| chr2_-_47572207 | 0.13 |
ENST00000441997.1
|
AC073283.4
|
AC073283.4 |
| chr11_-_67276100 | 0.13 |
ENST00000301488.3
|
CDK2AP2
|
cyclin-dependent kinase 2 associated protein 2 |
| chr20_-_17662878 | 0.13 |
ENST00000377813.1
ENST00000377807.2 ENST00000360807.4 ENST00000398782.2 |
RRBP1
|
ribosome binding protein 1 |
| chr19_-_10341948 | 0.13 |
ENST00000590320.1
ENST00000592342.1 ENST00000588952.1 |
S1PR2
DNMT1
|
sphingosine-1-phosphate receptor 2 DNA (cytosine-5-)-methyltransferase 1 |
| chr3_+_132378741 | 0.13 |
ENST00000493720.2
|
UBA5
|
ubiquitin-like modifier activating enzyme 5 |
| chrX_+_134478706 | 0.13 |
ENST00000370761.3
ENST00000339249.4 ENST00000370760.3 |
ZNF449
|
zinc finger protein 449 |
| chr5_+_94890778 | 0.13 |
ENST00000380009.4
|
ARSK
|
arylsulfatase family, member K |
| chr19_+_41256764 | 0.13 |
ENST00000243563.3
ENST00000601253.1 ENST00000597353.1 ENST00000599362.1 |
SNRPA
|
small nuclear ribonucleoprotein polypeptide A |
| chr7_-_91808845 | 0.13 |
ENST00000343318.5
|
LRRD1
|
leucine-rich repeats and death domain containing 1 |
| chr18_-_47017956 | 0.12 |
ENST00000584895.1
ENST00000332968.6 ENST00000580210.1 ENST00000579408.1 |
RPL17-C18orf32
RPL17
|
RPL17-C18orf32 readthrough ribosomal protein L17 |
| chr4_+_141445311 | 0.12 |
ENST00000323570.3
ENST00000511887.2 |
ELMOD2
|
ELMO/CED-12 domain containing 2 |
| chr6_+_24775641 | 0.12 |
ENST00000378054.2
ENST00000476555.1 |
GMNN
|
geminin, DNA replication inhibitor |
| chr16_+_2059872 | 0.12 |
ENST00000567649.1
|
NPW
|
neuropeptide W |
| chr5_+_65440032 | 0.12 |
ENST00000334121.6
|
SREK1
|
splicing regulatory glutamine/lysine-rich protein 1 |
| chr19_-_4670345 | 0.12 |
ENST00000599630.1
ENST00000262947.3 |
C19orf10
|
chromosome 19 open reading frame 10 |
| chr3_+_25824405 | 0.12 |
ENST00000452098.1
|
OXSM
|
3-oxoacyl-ACP synthase, mitochondrial |
| chr18_+_60190226 | 0.12 |
ENST00000269499.5
|
ZCCHC2
|
zinc finger, CCHC domain containing 2 |
| chr5_-_175965008 | 0.12 |
ENST00000537487.1
|
RNF44
|
ring finger protein 44 |
| chr2_+_183580954 | 0.11 |
ENST00000264065.7
|
DNAJC10
|
DnaJ (Hsp40) homolog, subfamily C, member 10 |
| chr9_+_139249228 | 0.11 |
ENST00000392944.1
|
GPSM1
|
G-protein signaling modulator 1 |
| chr19_+_39903185 | 0.11 |
ENST00000409794.3
|
PLEKHG2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr13_-_31040060 | 0.11 |
ENST00000326004.4
ENST00000341423.5 |
HMGB1
|
high mobility group box 1 |
| chrX_+_48660287 | 0.11 |
ENST00000444343.2
ENST00000376610.2 ENST00000334136.5 ENST00000376619.2 |
HDAC6
|
histone deacetylase 6 |
| chr16_-_57219721 | 0.11 |
ENST00000562406.1
ENST00000568671.1 ENST00000567044.1 |
FAM192A
|
family with sequence similarity 192, member A |
| chr1_+_1567474 | 0.11 |
ENST00000356026.5
|
MMP23B
|
matrix metallopeptidase 23B |
| chr17_-_77967433 | 0.11 |
ENST00000571872.1
|
TBC1D16
|
TBC1 domain family, member 16 |
| chr3_-_49395892 | 0.11 |
ENST00000419783.1
|
GPX1
|
glutathione peroxidase 1 |
| chrX_-_10544942 | 0.10 |
ENST00000380779.1
|
MID1
|
midline 1 (Opitz/BBB syndrome) |
| chr3_-_132378919 | 0.10 |
ENST00000355458.3
|
ACAD11
|
acyl-CoA dehydrogenase family, member 11 |
| chr16_-_73082274 | 0.10 |
ENST00000268489.5
|
ZFHX3
|
zinc finger homeobox 3 |
| chr2_-_114514181 | 0.10 |
ENST00000409342.1
|
SLC35F5
|
solute carrier family 35, member F5 |
| chr5_-_94890648 | 0.10 |
ENST00000513823.1
ENST00000514952.1 ENST00000358746.2 |
TTC37
|
tetratricopeptide repeat domain 37 |
| chr4_-_54930790 | 0.10 |
ENST00000263921.3
|
CHIC2
|
cysteine-rich hydrophobic domain 2 |
| chr2_+_192542850 | 0.10 |
ENST00000410026.2
|
NABP1
|
nucleic acid binding protein 1 |
| chr9_-_98079965 | 0.10 |
ENST00000289081.3
|
FANCC
|
Fanconi anemia, complementation group C |
| chr1_-_113498616 | 0.10 |
ENST00000433570.4
ENST00000538576.1 ENST00000458229.1 |
SLC16A1
|
solute carrier family 16 (monocarboxylate transporter), member 1 |
| chrX_-_153059958 | 0.10 |
ENST00000370092.3
ENST00000217901.5 |
IDH3G
|
isocitrate dehydrogenase 3 (NAD+) gamma |
| chr1_-_156399184 | 0.10 |
ENST00000368243.1
ENST00000357975.4 ENST00000310027.5 ENST00000400991.2 |
C1orf61
|
chromosome 1 open reading frame 61 |
| chr1_-_160313025 | 0.10 |
ENST00000368069.3
ENST00000241704.7 |
COPA
|
coatomer protein complex, subunit alpha |
| chr12_+_48722763 | 0.09 |
ENST00000335017.1
|
H1FNT
|
H1 histone family, member N, testis-specific |
| chr4_-_119757239 | 0.09 |
ENST00000280551.6
|
SEC24D
|
SEC24 family member D |
| chr8_-_29208183 | 0.09 |
ENST00000240100.2
|
DUSP4
|
dual specificity phosphatase 4 |
| chr16_-_57219966 | 0.09 |
ENST00000565760.1
ENST00000309137.8 ENST00000570184.1 ENST00000562324.1 |
FAM192A
|
family with sequence similarity 192, member A |
| chr13_-_36920420 | 0.09 |
ENST00000438666.2
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome) |
| chr17_-_46716647 | 0.09 |
ENST00000608940.1
|
RP11-357H14.17
|
RP11-357H14.17 |
| chr12_-_76953513 | 0.09 |
ENST00000547540.1
|
OSBPL8
|
oxysterol binding protein-like 8 |
| chr22_+_21271714 | 0.09 |
ENST00000354336.3
|
CRKL
|
v-crk avian sarcoma virus CT10 oncogene homolog-like |
| chr19_+_38810447 | 0.09 |
ENST00000263372.3
|
KCNK6
|
potassium channel, subfamily K, member 6 |
| chr7_+_128379449 | 0.09 |
ENST00000479257.1
|
CALU
|
calumenin |
| chr11_+_65686728 | 0.09 |
ENST00000312515.2
ENST00000525501.1 |
DRAP1
|
DR1-associated protein 1 (negative cofactor 2 alpha) |
| chr6_+_151561085 | 0.09 |
ENST00000402676.2
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr18_-_47018897 | 0.09 |
ENST00000418495.1
|
RPL17
|
ribosomal protein L17 |
| chr8_-_38126675 | 0.09 |
ENST00000531823.1
ENST00000534339.1 ENST00000524616.1 ENST00000422581.2 ENST00000424479.2 ENST00000419686.2 |
PPAPDC1B
|
phosphatidic acid phosphatase type 2 domain containing 1B |
| chr7_-_93633658 | 0.09 |
ENST00000433727.1
|
BET1
|
Bet1 golgi vesicular membrane trafficking protein |
| chr20_-_1306391 | 0.09 |
ENST00000339987.3
|
SDCBP2
|
syndecan binding protein (syntenin) 2 |
| chr19_+_797443 | 0.09 |
ENST00000394601.4
ENST00000589575.1 |
PTBP1
|
polypyrimidine tract binding protein 1 |
| chrX_+_48660107 | 0.09 |
ENST00000376643.2
|
HDAC6
|
histone deacetylase 6 |
| chr17_-_7218631 | 0.09 |
ENST00000577040.2
ENST00000389167.5 ENST00000391950.3 |
GPS2
|
G protein pathway suppressor 2 |
| chr16_+_19125252 | 0.09 |
ENST00000566735.1
ENST00000381440.3 |
ITPRIPL2
|
inositol 1,4,5-trisphosphate receptor interacting protein-like 2 |
| chr5_+_133984462 | 0.08 |
ENST00000398844.2
ENST00000322887.4 |
SEC24A
|
SEC24 family member A |
| chr13_-_36920872 | 0.08 |
ENST00000451493.1
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome) |
| chr19_-_5680891 | 0.08 |
ENST00000309324.4
|
C19orf70
|
chromosome 19 open reading frame 70 |
| chr6_+_24775153 | 0.08 |
ENST00000356509.3
ENST00000230056.3 |
GMNN
|
geminin, DNA replication inhibitor |
| chr1_+_160313165 | 0.08 |
ENST00000421914.1
ENST00000535857.1 ENST00000438008.1 |
NCSTN
|
nicastrin |
| chr19_-_40724246 | 0.08 |
ENST00000311308.6
|
TTC9B
|
tetratricopeptide repeat domain 9B |
| chr4_+_40058411 | 0.08 |
ENST00000261435.6
ENST00000515550.1 |
N4BP2
|
NEDD4 binding protein 2 |
| chr1_-_155145721 | 0.08 |
ENST00000295682.4
|
KRTCAP2
|
keratinocyte associated protein 2 |
| chr19_+_531713 | 0.08 |
ENST00000215574.4
|
CDC34
|
cell division cycle 34 |
| chr18_+_76829441 | 0.08 |
ENST00000458297.2
|
ATP9B
|
ATPase, class II, type 9B |
| chr7_+_128379346 | 0.08 |
ENST00000535011.2
ENST00000542996.2 ENST00000535623.1 ENST00000538546.1 ENST00000249364.4 ENST00000449187.2 |
CALU
|
calumenin |
| chr5_-_150080472 | 0.08 |
ENST00000521464.1
ENST00000518917.1 ENST00000447771.2 ENST00000540000.1 ENST00000199814.4 |
RBM22
|
RNA binding motif protein 22 |
| chr2_+_192542879 | 0.08 |
ENST00000409510.1
|
NABP1
|
nucleic acid binding protein 1 |
| chr17_+_6554971 | 0.08 |
ENST00000391428.2
|
C17orf100
|
chromosome 17 open reading frame 100 |
| chr14_+_31091511 | 0.08 |
ENST00000544052.2
ENST00000421551.3 ENST00000541123.1 ENST00000557076.1 ENST00000553693.1 ENST00000396629.2 |
SCFD1
|
sec1 family domain containing 1 |
| chr16_-_57219926 | 0.08 |
ENST00000566584.1
ENST00000566481.1 ENST00000566077.1 ENST00000564108.1 ENST00000565458.1 ENST00000566681.1 ENST00000567439.1 |
FAM192A
|
family with sequence similarity 192, member A |
| chr5_-_131563474 | 0.08 |
ENST00000417528.1
|
P4HA2
|
prolyl 4-hydroxylase, alpha polypeptide II |
| chrX_+_55744228 | 0.08 |
ENST00000262850.7
|
RRAGB
|
Ras-related GTP binding B |
| chr18_-_47018769 | 0.08 |
ENST00000583637.1
ENST00000578528.1 ENST00000578532.1 ENST00000580387.1 ENST00000579248.1 ENST00000581373.1 |
RPL17
|
ribosomal protein L17 |
| chr13_-_36920615 | 0.08 |
ENST00000494062.2
|
SPG20
|
spastic paraplegia 20 (Troyer syndrome) |
| chrX_+_10126488 | 0.08 |
ENST00000380829.1
ENST00000421085.2 ENST00000454850.1 |
CLCN4
|
chloride channel, voltage-sensitive 4 |
| chr6_+_30908747 | 0.08 |
ENST00000462446.1
ENST00000304311.2 |
DPCR1
|
diffuse panbronchiolitis critical region 1 |
| chr10_+_129785536 | 0.08 |
ENST00000419012.2
|
PTPRE
|
protein tyrosine phosphatase, receptor type, E |
| chr20_-_1306351 | 0.08 |
ENST00000381812.1
|
SDCBP2
|
syndecan binding protein (syntenin) 2 |
| chr5_-_39425222 | 0.08 |
ENST00000320816.6
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr1_+_60280458 | 0.07 |
ENST00000455990.1
ENST00000371208.3 |
HOOK1
|
hook microtubule-tethering protein 1 |
| chr17_-_73401567 | 0.07 |
ENST00000392562.1
|
GRB2
|
growth factor receptor-bound protein 2 |
| chr2_-_198299726 | 0.07 |
ENST00000409915.4
ENST00000487698.1 ENST00000414963.2 ENST00000335508.6 |
SF3B1
|
splicing factor 3b, subunit 1, 155kDa |
| chrX_+_55744166 | 0.07 |
ENST00000374941.4
ENST00000414239.1 |
RRAGB
|
Ras-related GTP binding B |
| chr7_+_100860949 | 0.07 |
ENST00000305105.2
|
ZNHIT1
|
zinc finger, HIT-type containing 1 |
| chr5_-_39425068 | 0.07 |
ENST00000515700.1
ENST00000339788.6 |
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr7_-_138720763 | 0.07 |
ENST00000275766.1
|
ZC3HAV1L
|
zinc finger CCCH-type, antiviral 1-like |
| chr22_+_46476192 | 0.07 |
ENST00000443490.1
|
FLJ27365
|
hsa-mir-4763 |
| chr16_+_68056844 | 0.07 |
ENST00000565263.1
|
DUS2
|
dihydrouridine synthase 2 |
| chr8_-_38126635 | 0.07 |
ENST00000529359.1
|
PPAPDC1B
|
phosphatidic acid phosphatase type 2 domain containing 1B |
| chr5_-_39425290 | 0.07 |
ENST00000545653.1
|
DAB2
|
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
| chr1_-_153940097 | 0.07 |
ENST00000413622.1
ENST00000310483.6 |
SLC39A1
|
solute carrier family 39 (zinc transporter), member 1 |
| chr11_+_65686802 | 0.07 |
ENST00000376991.2
|
DRAP1
|
DR1-associated protein 1 (negative cofactor 2 alpha) |
| chr1_+_200638629 | 0.07 |
ENST00000568695.1
|
RP11-92G12.3
|
RP11-92G12.3 |
| chrX_+_47441712 | 0.07 |
ENST00000218388.4
ENST00000377018.2 ENST00000456754.2 ENST00000377017.1 ENST00000441738.1 |
TIMP1
|
TIMP metallopeptidase inhibitor 1 |
| chr19_+_797392 | 0.07 |
ENST00000350092.4
ENST00000349038.4 ENST00000586481.1 ENST00000585535.1 |
PTBP1
|
polypyrimidine tract binding protein 1 |
| chr12_-_54121261 | 0.07 |
ENST00000549784.1
ENST00000262059.4 |
CALCOCO1
|
calcium binding and coiled-coil domain 1 |
| chr3_-_57583052 | 0.07 |
ENST00000496292.1
ENST00000489843.1 |
ARF4
|
ADP-ribosylation factor 4 |
| chr5_-_150138551 | 0.07 |
ENST00000446090.2
ENST00000447998.2 |
DCTN4
|
dynactin 4 (p62) |
| chr3_-_49395705 | 0.07 |
ENST00000419349.1
|
GPX1
|
glutathione peroxidase 1 |
| chr1_+_44445643 | 0.07 |
ENST00000309519.7
|
B4GALT2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr19_+_32836499 | 0.07 |
ENST00000311921.4
ENST00000544431.1 ENST00000355898.5 |
ZNF507
|
zinc finger protein 507 |
| chrX_-_153059811 | 0.07 |
ENST00000427365.2
ENST00000444450.1 ENST00000370093.1 |
IDH3G
|
isocitrate dehydrogenase 3 (NAD+) gamma |
| chr10_-_30348439 | 0.07 |
ENST00000375377.1
|
KIAA1462
|
KIAA1462 |
| chr1_+_11866207 | 0.06 |
ENST00000312413.6
ENST00000346436.6 |
CLCN6
|
chloride channel, voltage-sensitive 6 |
| chr17_+_79660798 | 0.06 |
ENST00000571237.1
|
HGS
|
hepatocyte growth factor-regulated tyrosine kinase substrate |
| chr12_-_113909877 | 0.06 |
ENST00000261731.3
|
LHX5
|
LIM homeobox 5 |
| chr22_+_39916558 | 0.06 |
ENST00000337304.2
ENST00000396680.1 |
ATF4
|
activating transcription factor 4 |
| chr5_-_131563501 | 0.06 |
ENST00000401867.1
ENST00000379086.1 ENST00000418055.1 ENST00000453286.1 ENST00000166534.4 |
P4HA2
|
prolyl 4-hydroxylase, alpha polypeptide II |
| chr3_+_121554046 | 0.06 |
ENST00000273668.2
ENST00000451944.2 |
EAF2
|
ELL associated factor 2 |
| chr10_-_90342947 | 0.06 |
ENST00000437752.1
ENST00000331772.4 |
RNLS
|
renalase, FAD-dependent amine oxidase |
| chr2_-_202645835 | 0.06 |
ENST00000264276.6
|
ALS2
|
amyotrophic lateral sclerosis 2 (juvenile) |
| chr6_+_36164487 | 0.06 |
ENST00000357641.6
|
BRPF3
|
bromodomain and PHD finger containing, 3 |
| chr8_+_110552831 | 0.06 |
ENST00000530629.1
|
EBAG9
|
estrogen receptor binding site associated, antigen, 9 |
| chr10_-_105437909 | 0.06 |
ENST00000540321.1
|
SH3PXD2A
|
SH3 and PX domains 2A |
| chr17_-_66951474 | 0.06 |
ENST00000269080.2
|
ABCA8
|
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr10_-_111683308 | 0.06 |
ENST00000502935.1
ENST00000322238.8 ENST00000369680.4 |
XPNPEP1
|
X-prolyl aminopeptidase (aminopeptidase P) 1, soluble |
Network of associatons between targets according to the STRING database.
First level regulatory network of CREB3L1_CREB3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:1990169 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.1 | 0.3 | GO:0015728 | mevalonate transport(GO:0015728) behavioral response to nutrient(GO:0051780) |
| 0.1 | 0.3 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.1 | 0.2 | GO:1903519 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.1 | 0.3 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.1 | 0.5 | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) |
| 0.1 | 0.3 | GO:0048698 | negative regulation of collateral sprouting in absence of injury(GO:0048698) |
| 0.1 | 0.3 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.1 | 0.2 | GO:0009443 | pyridoxal 5'-phosphate salvage(GO:0009443) |
| 0.0 | 0.2 | GO:0070845 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 0.0 | 0.2 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.2 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.0 | 0.2 | GO:0009609 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.0 | 0.3 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 0.0 | 0.2 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.0 | 0.5 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.2 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.2 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.0 | 0.1 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) regulation of restriction endodeoxyribonuclease activity(GO:0032072) negative regulation of apoptotic cell clearance(GO:2000426) |
| 0.0 | 0.3 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 0.0 | 0.2 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.3 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.5 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.2 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.2 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.1 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.0 | 0.1 | GO:0051792 | medium-chain fatty acid biosynthetic process(GO:0051792) |
| 0.0 | 0.2 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.2 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.0 | 0.1 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.0 | 0.1 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.0 | 0.2 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.1 | GO:0038178 | complement component C5a signaling pathway(GO:0038178) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.0 | 0.4 | GO:0035090 | maintenance of apical/basal cell polarity(GO:0035090) maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.1 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.1 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.4 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.1 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.0 | 0.3 | GO:0033182 | regulation of histone ubiquitination(GO:0033182) |
| 0.0 | 0.0 | GO:0045212 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.0 | 0.3 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.2 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.0 | 0.1 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.0 | 0.0 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.1 | 0.3 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.1 | 0.2 | GO:0008623 | CHRAC(GO:0008623) |
| 0.1 | 0.4 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.3 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.2 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.1 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.2 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.0 | 0.4 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.2 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.2 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.2 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.1 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.1 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.0 | 0.2 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.1 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.1 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.2 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.2 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 0.1 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.0 | 0.1 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0015130 | mevalonate transmembrane transporter activity(GO:0015130) |
| 0.1 | 0.2 | GO:0000224 | peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase activity(GO:0000224) |
| 0.1 | 0.4 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.1 | 0.2 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.1 | 0.2 | GO:0071566 | UFM1 activating enzyme activity(GO:0071566) |
| 0.1 | 0.2 | GO:0008478 | pyridoxal kinase activity(GO:0008478) |
| 0.0 | 0.2 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.2 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.6 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.0 | 0.1 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.0 | 0.1 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 0.0 | 0.4 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.2 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.0 | 0.3 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.2 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.2 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.2 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.2 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.0 | 0.1 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.3 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.1 | GO:0008441 | 3'(2'),5'-bisphosphate nucleotidase activity(GO:0008441) |
| 0.0 | 0.1 | GO:0004878 | complement component C5a receptor activity(GO:0004878) |
| 0.0 | 0.1 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.2 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.1 | GO:0010858 | calcium-dependent protein kinase regulator activity(GO:0010858) |
| 0.0 | 0.5 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.5 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.1 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.3 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.5 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.2 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |