Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
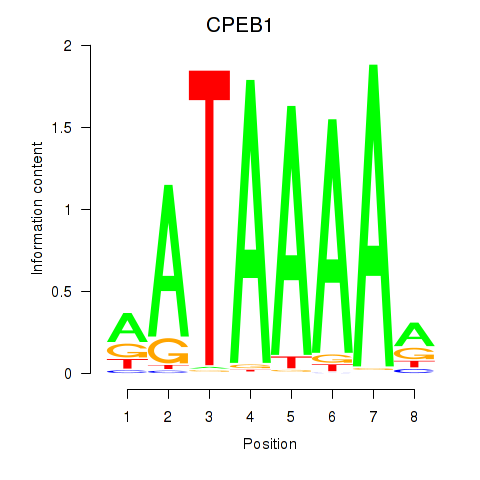
Results for CPEB1
Z-value: 0.52
Transcription factors associated with CPEB1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CPEB1
|
ENSG00000214575.5 | cytoplasmic polyadenylation element binding protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CPEB1 | hg19_v2_chr15_-_83240507_83240539 | 1.00 | 1.5e-03 | Click! |
Activity profile of CPEB1 motif
Sorted Z-values of CPEB1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_101733794 | 0.41 |
ENST00000523555.1
|
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr9_-_20622478 | 0.28 |
ENST00000355930.6
ENST00000380338.4 |
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr14_+_56078695 | 0.23 |
ENST00000416613.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr1_-_65533390 | 0.21 |
ENST00000448344.1
|
RP4-535B20.1
|
RP4-535B20.1 |
| chr1_+_116654376 | 0.19 |
ENST00000369500.3
|
MAB21L3
|
mab-21-like 3 (C. elegans) |
| chr21_-_35340759 | 0.19 |
ENST00000607953.1
|
AP000569.9
|
AP000569.9 |
| chr6_+_26021869 | 0.18 |
ENST00000359907.3
|
HIST1H4A
|
histone cluster 1, H4a |
| chr8_+_94752349 | 0.17 |
ENST00000391680.1
|
RBM12B-AS1
|
RBM12B antisense RNA 1 |
| chr2_-_55646412 | 0.16 |
ENST00000413716.2
|
CCDC88A
|
coiled-coil domain containing 88A |
| chr6_+_26104104 | 0.16 |
ENST00000377803.2
|
HIST1H4C
|
histone cluster 1, H4c |
| chr7_+_66800928 | 0.15 |
ENST00000430244.1
|
RP11-166O4.5
|
RP11-166O4.5 |
| chr12_-_12714006 | 0.14 |
ENST00000541207.1
|
DUSP16
|
dual specificity phosphatase 16 |
| chr1_-_11907829 | 0.14 |
ENST00000376480.3
|
NPPA
|
natriuretic peptide A |
| chr9_+_20927764 | 0.14 |
ENST00000603044.1
ENST00000604254.1 |
FOCAD
|
focadhesin |
| chr11_-_5248294 | 0.14 |
ENST00000335295.4
|
HBB
|
hemoglobin, beta |
| chr10_+_94590910 | 0.14 |
ENST00000371547.4
|
EXOC6
|
exocyst complex component 6 |
| chr9_-_20621834 | 0.14 |
ENST00000429426.2
|
MLLT3
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr4_-_65275162 | 0.13 |
ENST00000381210.3
ENST00000507440.1 |
TECRL
|
trans-2,3-enoyl-CoA reductase-like |
| chrX_-_138914394 | 0.13 |
ENST00000327569.3
ENST00000361648.2 ENST00000370543.1 ENST00000359686.2 |
ATP11C
|
ATPase, class VI, type 11C |
| chr2_-_190927447 | 0.13 |
ENST00000260950.4
|
MSTN
|
myostatin |
| chr2_-_190044480 | 0.13 |
ENST00000374866.3
|
COL5A2
|
collagen, type V, alpha 2 |
| chr1_+_212475148 | 0.12 |
ENST00000537030.3
|
PPP2R5A
|
protein phosphatase 2, regulatory subunit B', alpha |
| chr12_+_41221794 | 0.12 |
ENST00000547849.1
|
CNTN1
|
contactin 1 |
| chr1_-_11918988 | 0.12 |
ENST00000376468.3
|
NPPB
|
natriuretic peptide B |
| chr6_+_15401075 | 0.11 |
ENST00000541660.1
|
JARID2
|
jumonji, AT rich interactive domain 2 |
| chr5_+_139505520 | 0.11 |
ENST00000333305.3
|
IGIP
|
IgA-inducing protein |
| chr14_-_69444957 | 0.11 |
ENST00000556571.1
ENST00000553659.1 ENST00000555616.1 |
ACTN1
|
actinin, alpha 1 |
| chr3_+_160394940 | 0.11 |
ENST00000320767.2
|
ARL14
|
ADP-ribosylation factor-like 14 |
| chr4_-_102267953 | 0.10 |
ENST00000523694.2
ENST00000507176.1 |
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr3_+_107241783 | 0.10 |
ENST00000415149.2
ENST00000402543.1 ENST00000325805.8 ENST00000427402.1 |
BBX
|
bobby sox homolog (Drosophila) |
| chr3_-_148939598 | 0.10 |
ENST00000455472.3
|
CP
|
ceruloplasmin (ferroxidase) |
| chr10_-_10504285 | 0.10 |
ENST00000602311.1
|
RP11-271F18.4
|
RP11-271F18.4 |
| chr3_+_41236325 | 0.10 |
ENST00000426215.1
ENST00000405570.1 |
CTNNB1
|
catenin (cadherin-associated protein), beta 1, 88kDa |
| chr20_-_10654639 | 0.10 |
ENST00000254958.5
|
JAG1
|
jagged 1 |
| chr1_+_205682497 | 0.10 |
ENST00000598338.1
|
AC119673.1
|
AC119673.1 |
| chr5_+_131993856 | 0.10 |
ENST00000304506.3
|
IL13
|
interleukin 13 |
| chr17_-_38574169 | 0.10 |
ENST00000423485.1
|
TOP2A
|
topoisomerase (DNA) II alpha 170kDa |
| chr6_+_111195973 | 0.10 |
ENST00000368885.3
ENST00000368882.3 ENST00000451850.2 ENST00000368877.5 |
AMD1
|
adenosylmethionine decarboxylase 1 |
| chrX_-_20236970 | 0.10 |
ENST00000379548.4
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| chr6_+_27100811 | 0.09 |
ENST00000359193.2
|
HIST1H2AG
|
histone cluster 1, H2ag |
| chr8_+_26435915 | 0.09 |
ENST00000523027.1
|
DPYSL2
|
dihydropyrimidinase-like 2 |
| chr8_+_81397846 | 0.09 |
ENST00000379091.4
|
ZBTB10
|
zinc finger and BTB domain containing 10 |
| chr2_+_64073187 | 0.09 |
ENST00000491621.1
|
UGP2
|
UDP-glucose pyrophosphorylase 2 |
| chr7_-_20826504 | 0.09 |
ENST00000418710.2
ENST00000361443.4 |
SP8
|
Sp8 transcription factor |
| chr11_-_47521309 | 0.09 |
ENST00000535982.1
|
CELF1
|
CUGBP, Elav-like family member 1 |
| chr4_-_76957214 | 0.09 |
ENST00000306621.3
|
CXCL11
|
chemokine (C-X-C motif) ligand 11 |
| chr2_+_181845763 | 0.09 |
ENST00000602499.1
|
UBE2E3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr7_-_27219849 | 0.09 |
ENST00000396344.4
|
HOXA10
|
homeobox A10 |
| chr1_-_32384693 | 0.09 |
ENST00000602683.1
ENST00000470404.1 |
PTP4A2
|
protein tyrosine phosphatase type IVA, member 2 |
| chr2_-_206950996 | 0.09 |
ENST00000414320.1
|
INO80D
|
INO80 complex subunit D |
| chr1_+_41447609 | 0.08 |
ENST00000543104.1
|
CTPS1
|
CTP synthase 1 |
| chr9_+_22646189 | 0.08 |
ENST00000436786.1
|
RP11-399D6.2
|
RP11-399D6.2 |
| chr2_-_207023918 | 0.08 |
ENST00000455934.2
ENST00000449699.1 ENST00000454195.1 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr11_-_11374904 | 0.08 |
ENST00000528848.2
|
CSNK2A3
|
casein kinase 2, alpha 3 polypeptide |
| chr9_-_16253112 | 0.08 |
ENST00000380683.1
|
C9orf92
|
chromosome 9 open reading frame 92 |
| chr8_-_95449155 | 0.08 |
ENST00000481490.2
|
FSBP
|
fibrinogen silencer binding protein |
| chr1_-_200379180 | 0.08 |
ENST00000294740.3
|
ZNF281
|
zinc finger protein 281 |
| chr13_-_46626847 | 0.08 |
ENST00000242848.4
ENST00000282007.3 |
ZC3H13
|
zinc finger CCCH-type containing 13 |
| chr2_+_152214098 | 0.08 |
ENST00000243347.3
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr9_+_97562440 | 0.08 |
ENST00000395357.2
|
C9orf3
|
chromosome 9 open reading frame 3 |
| chr1_+_84630053 | 0.08 |
ENST00000394838.2
ENST00000370682.3 ENST00000432111.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr4_-_41884620 | 0.08 |
ENST00000504870.1
|
LINC00682
|
long intergenic non-protein coding RNA 682 |
| chr2_-_151344172 | 0.08 |
ENST00000375734.2
ENST00000263895.4 ENST00000454202.1 |
RND3
|
Rho family GTPase 3 |
| chr5_+_110073853 | 0.08 |
ENST00000513807.1
ENST00000509442.2 |
SLC25A46
|
solute carrier family 25, member 46 |
| chr17_-_39041479 | 0.07 |
ENST00000167588.3
|
KRT20
|
keratin 20 |
| chr12_-_96184913 | 0.07 |
ENST00000538383.1
|
NTN4
|
netrin 4 |
| chr4_-_40632140 | 0.07 |
ENST00000514782.1
|
RBM47
|
RNA binding motif protein 47 |
| chr12_-_49412198 | 0.07 |
ENST00000552463.1
|
PRKAG1
|
protein kinase, AMP-activated, gamma 1 non-catalytic subunit |
| chr11_+_77300669 | 0.07 |
ENST00000313578.3
|
AQP11
|
aquaporin 11 |
| chr2_+_88047606 | 0.07 |
ENST00000359481.4
|
PLGLB2
|
plasminogen-like B2 |
| chr4_+_174089951 | 0.07 |
ENST00000512285.1
|
GALNT7
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7) |
| chr12_-_92536433 | 0.07 |
ENST00000551563.2
ENST00000546975.1 ENST00000549802.1 |
C12orf79
|
chromosome 12 open reading frame 79 |
| chr1_+_82266053 | 0.07 |
ENST00000370715.1
ENST00000370713.1 ENST00000319517.6 ENST00000370717.2 ENST00000394879.1 ENST00000271029.4 ENST00000335786.5 |
LPHN2
|
latrophilin 2 |
| chr18_-_33702078 | 0.07 |
ENST00000586829.1
|
SLC39A6
|
solute carrier family 39 (zinc transporter), member 6 |
| chr3_-_196911002 | 0.07 |
ENST00000452595.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr8_-_95220775 | 0.07 |
ENST00000441892.2
ENST00000521491.1 ENST00000027335.3 |
CDH17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr14_-_62217779 | 0.07 |
ENST00000554254.1
|
HIF1A-AS2
|
HIF1A antisense RNA 2 |
| chr14_+_56127960 | 0.07 |
ENST00000553624.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr15_+_93426514 | 0.07 |
ENST00000556722.1
|
CHD2
|
chromodomain helicase DNA binding protein 2 |
| chr12_-_49412525 | 0.07 |
ENST00000551121.1
ENST00000552212.1 ENST00000548605.1 ENST00000548950.1 ENST00000547125.1 |
PRKAG1
|
protein kinase, AMP-activated, gamma 1 non-catalytic subunit |
| chr11_+_17281900 | 0.07 |
ENST00000530527.1
|
NUCB2
|
nucleobindin 2 |
| chr2_-_55277654 | 0.07 |
ENST00000337526.6
ENST00000317610.7 ENST00000357732.4 |
RTN4
|
reticulon 4 |
| chr6_+_26124373 | 0.07 |
ENST00000377791.2
ENST00000602637.1 |
HIST1H2AC
|
histone cluster 1, H2ac |
| chr11_+_123325106 | 0.07 |
ENST00000525757.1
|
LINC01059
|
long intergenic non-protein coding RNA 1059 |
| chr5_+_56471592 | 0.07 |
ENST00000511209.1
|
GPBP1
|
GC-rich promoter binding protein 1 |
| chr14_+_56127989 | 0.07 |
ENST00000555573.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr14_-_102552659 | 0.06 |
ENST00000441629.2
|
HSP90AA1
|
heat shock protein 90kDa alpha (cytosolic), class A member 1 |
| chr12_-_118797475 | 0.06 |
ENST00000541786.1
ENST00000419821.2 ENST00000541878.1 |
TAOK3
|
TAO kinase 3 |
| chr12_+_79258444 | 0.06 |
ENST00000261205.4
|
SYT1
|
synaptotagmin I |
| chr16_+_53133070 | 0.06 |
ENST00000565832.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr7_+_90012986 | 0.06 |
ENST00000416322.1
|
CLDN12
|
claudin 12 |
| chr20_-_40247002 | 0.06 |
ENST00000373222.3
|
CHD6
|
chromodomain helicase DNA binding protein 6 |
| chr6_+_31683117 | 0.06 |
ENST00000375825.3
ENST00000375824.1 |
LY6G6D
|
lymphocyte antigen 6 complex, locus G6D |
| chr5_+_162932554 | 0.06 |
ENST00000321757.6
ENST00000421814.2 ENST00000518095.1 |
MAT2B
|
methionine adenosyltransferase II, beta |
| chr2_-_175711133 | 0.06 |
ENST00000409597.1
ENST00000413882.1 |
CHN1
|
chimerin 1 |
| chr9_-_119162885 | 0.06 |
ENST00000445861.2
|
PAPPA-AS1
|
PAPPA antisense RNA 1 |
| chr12_+_28410128 | 0.06 |
ENST00000381259.1
ENST00000381256.1 |
CCDC91
|
coiled-coil domain containing 91 |
| chr2_-_158295915 | 0.06 |
ENST00000418920.1
|
CYTIP
|
cytohesin 1 interacting protein |
| chr11_-_102651343 | 0.06 |
ENST00000279441.4
ENST00000539681.1 |
MMP10
|
matrix metallopeptidase 10 (stromelysin 2) |
| chr9_+_42671887 | 0.06 |
ENST00000456520.1
ENST00000377391.3 |
CBWD7
|
COBW domain containing 7 |
| chr8_+_104831472 | 0.06 |
ENST00000262231.10
ENST00000507740.1 |
RIMS2
|
regulating synaptic membrane exocytosis 2 |
| chr4_-_102268484 | 0.06 |
ENST00000394853.4
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr12_+_79258547 | 0.06 |
ENST00000457153.2
|
SYT1
|
synaptotagmin I |
| chr1_+_84767289 | 0.06 |
ENST00000394834.3
ENST00000370669.1 |
SAMD13
|
sterile alpha motif domain containing 13 |
| chr4_-_105416039 | 0.06 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr6_+_117002339 | 0.06 |
ENST00000413340.1
ENST00000368564.1 ENST00000356348.1 |
KPNA5
|
karyopherin alpha 5 (importin alpha 6) |
| chr1_-_1763721 | 0.06 |
ENST00000437146.1
|
GNB1
|
guanine nucleotide binding protein (G protein), beta polypeptide 1 |
| chr13_-_67802549 | 0.06 |
ENST00000328454.5
ENST00000377865.2 |
PCDH9
|
protocadherin 9 |
| chr18_+_3252206 | 0.06 |
ENST00000578562.2
|
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr20_+_58179582 | 0.06 |
ENST00000371015.1
ENST00000395639.4 |
PHACTR3
|
phosphatase and actin regulator 3 |
| chrX_-_20237059 | 0.06 |
ENST00000457145.1
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| chr7_+_16700806 | 0.06 |
ENST00000446596.1
ENST00000438834.1 |
BZW2
|
basic leucine zipper and W2 domains 2 |
| chr12_-_52761262 | 0.05 |
ENST00000257901.3
|
KRT85
|
keratin 85 |
| chr1_+_161736072 | 0.05 |
ENST00000367942.3
|
ATF6
|
activating transcription factor 6 |
| chr12_-_50616382 | 0.05 |
ENST00000552783.1
|
LIMA1
|
LIM domain and actin binding 1 |
| chr6_-_34639733 | 0.05 |
ENST00000374021.1
|
C6orf106
|
chromosome 6 open reading frame 106 |
| chr1_-_152552980 | 0.05 |
ENST00000368787.3
|
LCE3D
|
late cornified envelope 3D |
| chr11_-_115127611 | 0.05 |
ENST00000545094.1
|
CADM1
|
cell adhesion molecule 1 |
| chr3_-_148939835 | 0.05 |
ENST00000264613.6
|
CP
|
ceruloplasmin (ferroxidase) |
| chr4_-_52883786 | 0.05 |
ENST00000343457.3
|
LRRC66
|
leucine rich repeat containing 66 |
| chr3_-_112360116 | 0.05 |
ENST00000206423.3
ENST00000439685.2 |
CCDC80
|
coiled-coil domain containing 80 |
| chr7_-_111846435 | 0.05 |
ENST00000437633.1
ENST00000428084.1 |
DOCK4
|
dedicator of cytokinesis 4 |
| chr8_-_95274536 | 0.05 |
ENST00000297596.2
ENST00000396194.2 |
GEM
|
GTP binding protein overexpressed in skeletal muscle |
| chr10_-_74856608 | 0.05 |
ENST00000307116.2
ENST00000373008.2 ENST00000412021.2 ENST00000394890.2 ENST00000263556.3 ENST00000440381.1 |
P4HA1
|
prolyl 4-hydroxylase, alpha polypeptide I |
| chr11_+_126276655 | 0.05 |
ENST00000524860.1
|
ST3GAL4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr18_-_33709268 | 0.05 |
ENST00000269187.5
ENST00000590986.1 ENST00000440549.2 |
SLC39A6
|
solute carrier family 39 (zinc transporter), member 6 |
| chr13_+_49684445 | 0.05 |
ENST00000398316.3
|
FNDC3A
|
fibronectin type III domain containing 3A |
| chrX_+_106871713 | 0.05 |
ENST00000372435.4
ENST00000372428.4 ENST00000372419.3 ENST00000543248.1 |
PRPS1
|
phosphoribosyl pyrophosphate synthetase 1 |
| chr4_-_52904425 | 0.05 |
ENST00000535450.1
|
SGCB
|
sarcoglycan, beta (43kDa dystrophin-associated glycoprotein) |
| chr6_-_27100529 | 0.05 |
ENST00000607124.1
ENST00000339812.2 ENST00000541790.1 |
HIST1H2BJ
|
histone cluster 1, H2bj |
| chr15_-_76304731 | 0.05 |
ENST00000394907.3
|
NRG4
|
neuregulin 4 |
| chr5_+_32788945 | 0.05 |
ENST00000326958.1
|
AC026703.1
|
AC026703.1 |
| chrX_+_154299690 | 0.05 |
ENST00000340647.4
ENST00000330045.7 |
BRCC3
|
BRCA1/BRCA2-containing complex, subunit 3 |
| chr10_-_21806759 | 0.05 |
ENST00000444772.3
|
SKIDA1
|
SKI/DACH domain containing 1 |
| chr7_-_104909435 | 0.05 |
ENST00000357311.3
|
SRPK2
|
SRSF protein kinase 2 |
| chr1_+_23037323 | 0.05 |
ENST00000544305.1
ENST00000374630.3 ENST00000400191.3 ENST00000374632.3 |
EPHB2
|
EPH receptor B2 |
| chr6_-_10115007 | 0.05 |
ENST00000485268.1
|
OFCC1
|
orofacial cleft 1 candidate 1 |
| chr17_-_73775839 | 0.05 |
ENST00000592643.1
ENST00000591890.1 ENST00000587171.1 ENST00000254810.4 ENST00000589599.1 |
H3F3B
|
H3 histone, family 3B (H3.3B) |
| chr8_-_90993869 | 0.05 |
ENST00000517772.1
|
NBN
|
nibrin |
| chr3_+_99357319 | 0.05 |
ENST00000452013.1
ENST00000261037.3 ENST00000273342.4 |
COL8A1
|
collagen, type VIII, alpha 1 |
| chr10_+_5406935 | 0.05 |
ENST00000380433.3
|
UCN3
|
urocortin 3 |
| chr1_+_61869748 | 0.05 |
ENST00000357977.5
|
NFIA
|
nuclear factor I/A |
| chr7_+_100464760 | 0.05 |
ENST00000200457.4
|
TRIP6
|
thyroid hormone receptor interactor 6 |
| chr12_-_125002827 | 0.05 |
ENST00000420698.1
|
NCOR2
|
nuclear receptor corepressor 2 |
| chr4_+_39046615 | 0.05 |
ENST00000261425.3
ENST00000508137.2 |
KLHL5
|
kelch-like family member 5 |
| chr10_+_121578211 | 0.05 |
ENST00000369080.3
|
INPP5F
|
inositol polyphosphate-5-phosphatase F |
| chr7_-_14029283 | 0.05 |
ENST00000433547.1
ENST00000405192.2 |
ETV1
|
ets variant 1 |
| chr3_-_93747425 | 0.05 |
ENST00000315099.2
|
STX19
|
syntaxin 19 |
| chr15_-_55562479 | 0.05 |
ENST00000564609.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr17_+_47075023 | 0.05 |
ENST00000431824.2
|
IGF2BP1
|
insulin-like growth factor 2 mRNA binding protein 1 |
| chr6_+_44194762 | 0.04 |
ENST00000371708.1
|
SLC29A1
|
solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
| chrX_+_73164149 | 0.04 |
ENST00000602938.1
ENST00000602294.1 ENST00000602920.1 ENST00000602737.1 ENST00000602772.1 |
JPX
|
JPX transcript, XIST activator (non-protein coding) |
| chrX_+_135279179 | 0.04 |
ENST00000370676.3
|
FHL1
|
four and a half LIM domains 1 |
| chr18_+_68002675 | 0.04 |
ENST00000584919.1
|
RP11-41O4.1
|
Uncharacterized protein |
| chr4_+_174089904 | 0.04 |
ENST00000265000.4
|
GALNT7
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7) |
| chr7_-_33080506 | 0.04 |
ENST00000381626.2
ENST00000409467.1 ENST00000449201.1 |
NT5C3A
|
5'-nucleotidase, cytosolic IIIA |
| chr9_-_215744 | 0.04 |
ENST00000382387.2
|
C9orf66
|
chromosome 9 open reading frame 66 |
| chr17_+_7482785 | 0.04 |
ENST00000250092.6
ENST00000380498.6 ENST00000584502.1 |
CD68
|
CD68 molecule |
| chr10_-_13523073 | 0.04 |
ENST00000440282.1
|
BEND7
|
BEN domain containing 7 |
| chr19_-_58485895 | 0.04 |
ENST00000314391.3
|
C19orf18
|
chromosome 19 open reading frame 18 |
| chr17_+_7533439 | 0.04 |
ENST00000441599.2
ENST00000380450.4 ENST00000416273.3 ENST00000575903.1 ENST00000576830.1 ENST00000571153.1 ENST00000575618.1 ENST00000576152.1 |
SHBG
|
sex hormone-binding globulin |
| chr3_-_69129501 | 0.04 |
ENST00000540295.1
ENST00000415609.2 ENST00000361055.4 ENST00000349511.4 |
UBA3
|
ubiquitin-like modifier activating enzyme 3 |
| chr3_-_196910721 | 0.04 |
ENST00000443183.1
|
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr17_-_39646116 | 0.04 |
ENST00000328119.6
|
KRT36
|
keratin 36 |
| chr2_+_173292390 | 0.04 |
ENST00000442250.1
ENST00000458358.1 ENST00000409080.1 |
ITGA6
|
integrin, alpha 6 |
| chr1_+_164529004 | 0.04 |
ENST00000559240.1
ENST00000367897.1 ENST00000540236.1 ENST00000401534.1 |
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr1_+_84629976 | 0.04 |
ENST00000446538.1
ENST00000370684.1 ENST00000436133.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr7_+_35756186 | 0.04 |
ENST00000430518.1
|
AC018647.3
|
AC018647.3 |
| chr1_+_169077133 | 0.04 |
ENST00000494797.1
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr4_+_170581213 | 0.04 |
ENST00000507875.1
|
CLCN3
|
chloride channel, voltage-sensitive 3 |
| chr17_-_1029052 | 0.04 |
ENST00000574437.1
|
ABR
|
active BCR-related |
| chr9_-_70465758 | 0.04 |
ENST00000489273.1
|
CBWD5
|
COBW domain containing 5 |
| chr1_+_52682052 | 0.04 |
ENST00000371591.1
|
ZFYVE9
|
zinc finger, FYVE domain containing 9 |
| chr14_+_39703112 | 0.04 |
ENST00000555143.1
ENST00000280082.3 |
MIA2
|
melanoma inhibitory activity 2 |
| chr4_+_95972822 | 0.04 |
ENST00000509540.1
ENST00000440890.2 |
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr7_-_111424506 | 0.04 |
ENST00000450156.1
ENST00000494651.2 |
DOCK4
|
dedicator of cytokinesis 4 |
| chr10_-_104262460 | 0.04 |
ENST00000446605.2
ENST00000369905.4 ENST00000545684.1 |
ACTR1A
|
ARP1 actin-related protein 1 homolog A, centractin alpha (yeast) |
| chr5_-_78808617 | 0.04 |
ENST00000282260.6
ENST00000508576.1 ENST00000535690.1 |
HOMER1
|
homer homolog 1 (Drosophila) |
| chr1_-_219615984 | 0.04 |
ENST00000420762.1
|
RP11-95P13.1
|
RP11-95P13.1 |
| chr7_-_14029515 | 0.04 |
ENST00000430479.1
ENST00000405218.2 ENST00000343495.5 |
ETV1
|
ets variant 1 |
| chr7_+_7811992 | 0.04 |
ENST00000406829.1
|
RPA3-AS1
|
RPA3 antisense RNA 1 |
| chr9_+_82187487 | 0.04 |
ENST00000435650.1
ENST00000414465.1 ENST00000376537.4 ENST00000376534.4 |
TLE4
|
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr1_+_164528866 | 0.04 |
ENST00000420696.2
|
PBX1
|
pre-B-cell leukemia homeobox 1 |
| chr18_+_3252265 | 0.04 |
ENST00000580887.1
ENST00000536605.1 |
MYL12A
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr5_-_73937244 | 0.04 |
ENST00000302351.4
ENST00000510316.1 ENST00000508331.1 |
ENC1
|
ectodermal-neural cortex 1 (with BTB domain) |
| chr17_-_15244894 | 0.04 |
ENST00000338696.2
ENST00000543896.1 ENST00000539245.1 ENST00000539316.1 ENST00000395930.1 |
TEKT3
|
tektin 3 |
| chr4_+_184365744 | 0.04 |
ENST00000504169.1
ENST00000302350.4 |
CDKN2AIP
|
CDKN2A interacting protein |
| chr10_+_47894023 | 0.04 |
ENST00000358474.5
|
FAM21B
|
family with sequence similarity 21, member B |
| chr4_+_184365841 | 0.04 |
ENST00000510928.1
|
CDKN2AIP
|
CDKN2A interacting protein |
| chr2_+_192543694 | 0.04 |
ENST00000435931.1
|
NABP1
|
nucleic acid binding protein 1 |
| chr14_-_35344093 | 0.04 |
ENST00000382422.2
|
BAZ1A
|
bromodomain adjacent to zinc finger domain, 1A |
| chr1_-_9129735 | 0.04 |
ENST00000377424.4
|
SLC2A5
|
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr2_+_183582774 | 0.04 |
ENST00000537515.1
|
DNAJC10
|
DnaJ (Hsp40) homolog, subfamily C, member 10 |
| chr17_+_44701402 | 0.04 |
ENST00000575068.1
|
NSF
|
N-ethylmaleimide-sensitive factor |
| chr12_-_109797249 | 0.04 |
ENST00000538041.1
|
RP11-256L11.1
|
RP11-256L11.1 |
| chr5_+_96077888 | 0.04 |
ENST00000509259.1
ENST00000503828.1 |
CAST
|
calpastatin |
| chr17_+_39846114 | 0.04 |
ENST00000586699.1
|
EIF1
|
eukaryotic translation initiation factor 1 |
| chr1_-_12679171 | 0.04 |
ENST00000606790.1
|
RP11-474O21.5
|
RP11-474O21.5 |
| chr4_-_74864386 | 0.04 |
ENST00000296027.4
|
CXCL5
|
chemokine (C-X-C motif) ligand 5 |
| chr1_+_169077172 | 0.04 |
ENST00000499679.3
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide |
Network of associatons between targets according to the STRING database.
First level regulatory network of CPEB1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.2 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.2 | GO:0045209 | MAPK phosphatase export from nucleus(GO:0045208) MAPK phosphatase export from nucleus, leptomycin B sensitive(GO:0045209) |
| 0.0 | 0.4 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.1 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.0 | 0.1 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.0 | 0.1 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.1 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.0 | 0.1 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 0.0 | 0.1 | GO:0061324 | glial cell fate determination(GO:0007403) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of chromatin-mediated maintenance of transcription(GO:1904499) positive regulation of chromatin-mediated maintenance of transcription(GO:1904501) regulation of euchromatin binding(GO:1904793) |
| 0.0 | 0.1 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.0 | 0.0 | GO:0051389 | inactivation of MAPKK activity(GO:0051389) |
| 0.0 | 0.4 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.1 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 0.2 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.0 | 0.0 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.0 | 0.1 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.0 | 0.1 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 | 0.1 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.1 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.1 | GO:0046101 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.0 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.2 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.1 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.1 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.0 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.0 | 0.0 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.1 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.2 | GO:1903286 | regulation of potassium ion import(GO:1903286) |
| 0.0 | 0.1 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.1 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.2 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.1 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.0 | 0.1 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.1 | GO:0034676 | integrin alpha6-beta4 complex(GO:0034676) |
| 0.0 | 0.1 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.1 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.1 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.0 | GO:0008623 | CHRAC(GO:0008623) |
| 0.0 | 0.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.1 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.1 | GO:0098642 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.1 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.1 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.1 | GO:0005427 | proton-dependent oligopeptide secondary active transmembrane transporter activity(GO:0005427) secondary active oligopeptide transmembrane transporter activity(GO:0015322) |
| 0.0 | 0.4 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.1 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.0 | GO:0019781 | NEDD8 activating enzyme activity(GO:0019781) |
| 0.0 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.2 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0051748 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.0 | 0.0 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID AP1 PATHWAY | AP-1 transcription factor network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |