Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
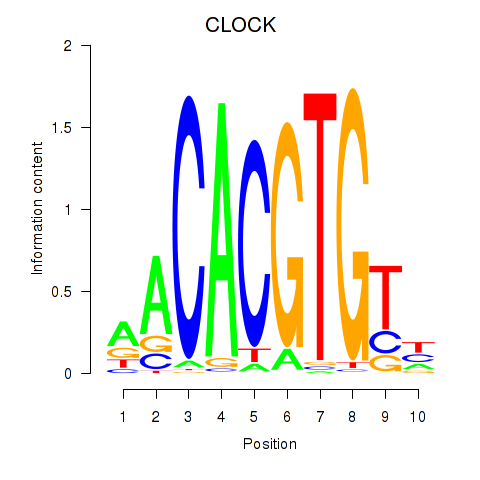
Results for CLOCK
Z-value: 0.54
Transcription factors associated with CLOCK
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CLOCK
|
ENSG00000134852.10 | clock circadian regulator |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CLOCK | hg19_v2_chr4_-_56412713_56412799 | -0.23 | 7.7e-01 | Click! |
Activity profile of CLOCK motif
Sorted Z-values of CLOCK motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_3704822 | 0.28 |
ENST00000414110.2
|
DNASE1
|
deoxyribonuclease I |
| chr3_+_72201910 | 0.27 |
ENST00000469178.1
ENST00000485404.1 |
LINC00870
|
long intergenic non-protein coding RNA 870 |
| chr4_+_17578815 | 0.25 |
ENST00000226299.4
|
LAP3
|
leucine aminopeptidase 3 |
| chr4_+_17579110 | 0.22 |
ENST00000606142.1
|
LAP3
|
leucine aminopeptidase 3 |
| chr4_+_183065793 | 0.19 |
ENST00000512480.1
|
TENM3
|
teneurin transmembrane protein 3 |
| chr22_-_50968419 | 0.19 |
ENST00000425169.1
ENST00000395680.1 ENST00000395681.1 ENST00000395678.3 ENST00000252029.3 |
TYMP
|
thymidine phosphorylase |
| chr5_-_172755056 | 0.19 |
ENST00000520648.1
|
STC2
|
stanniocalcin 2 |
| chr7_+_105172612 | 0.18 |
ENST00000493041.1
|
RINT1
|
RAD50 interactor 1 |
| chr16_+_66461175 | 0.17 |
ENST00000536005.2
ENST00000299694.8 ENST00000561796.1 |
BEAN1
|
brain expressed, associated with NEDD4, 1 |
| chr18_-_2982869 | 0.16 |
ENST00000584915.1
|
LPIN2
|
lipin 2 |
| chr2_+_131769256 | 0.14 |
ENST00000355771.3
|
ARHGEF4
|
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr6_+_149539767 | 0.14 |
ENST00000606202.1
ENST00000536230.1 ENST00000445901.1 |
TAB2
RP1-111D6.3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 2 RP1-111D6.3 |
| chr5_+_34656529 | 0.14 |
ENST00000513974.1
ENST00000512629.1 |
RAI14
|
retinoic acid induced 14 |
| chr6_+_32811861 | 0.14 |
ENST00000453426.1
|
TAPSAR1
|
TAP1 and PSMB8 antisense RNA 1 |
| chr7_-_92465868 | 0.13 |
ENST00000424848.2
|
CDK6
|
cyclin-dependent kinase 6 |
| chr7_-_130597935 | 0.13 |
ENST00000447307.1
ENST00000418546.1 |
MIR29B1
|
microRNA 29a |
| chr14_-_68066849 | 0.13 |
ENST00000558493.1
ENST00000561272.1 |
PIGH
|
phosphatidylinositol glycan anchor biosynthesis, class H |
| chr2_+_66918558 | 0.13 |
ENST00000435389.1
ENST00000428590.1 ENST00000412944.1 |
AC007392.3
|
AC007392.3 |
| chr19_-_42759266 | 0.13 |
ENST00000594664.1
|
AC006486.9
|
Uncharacterized protein |
| chr11_+_72929319 | 0.13 |
ENST00000393597.2
ENST00000311131.2 |
P2RY2
|
purinergic receptor P2Y, G-protein coupled, 2 |
| chr12_+_57623907 | 0.12 |
ENST00000553529.1
ENST00000554310.1 |
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr5_+_135394840 | 0.12 |
ENST00000503087.1
|
TGFBI
|
transforming growth factor, beta-induced, 68kDa |
| chr10_-_126849626 | 0.12 |
ENST00000530884.1
|
CTBP2
|
C-terminal binding protein 2 |
| chr4_+_139936905 | 0.12 |
ENST00000280614.2
|
CCRN4L
|
CCR4 carbon catabolite repression 4-like (S. cerevisiae) |
| chr5_+_34656450 | 0.12 |
ENST00000514527.1
|
RAI14
|
retinoic acid induced 14 |
| chr5_+_34656569 | 0.11 |
ENST00000428746.2
|
RAI14
|
retinoic acid induced 14 |
| chr3_-_52719888 | 0.11 |
ENST00000458294.1
|
PBRM1
|
polybromo 1 |
| chr10_+_60028818 | 0.11 |
ENST00000333926.5
|
CISD1
|
CDGSH iron sulfur domain 1 |
| chr7_-_1781906 | 0.11 |
ENST00000453348.1
ENST00000415399.1 |
AC074389.9
|
AC074389.9 |
| chr1_+_84944926 | 0.11 |
ENST00000370656.1
ENST00000370654.5 |
RPF1
|
ribosome production factor 1 homolog (S. cerevisiae) |
| chr12_-_46766577 | 0.11 |
ENST00000256689.5
|
SLC38A2
|
solute carrier family 38, member 2 |
| chr8_+_95908041 | 0.10 |
ENST00000396113.1
ENST00000519136.1 |
NDUFAF6
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr12_+_57624059 | 0.10 |
ENST00000557427.1
|
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr4_-_1107306 | 0.10 |
ENST00000433731.2
ENST00000333673.5 ENST00000382968.5 |
RNF212
|
ring finger protein 212 |
| chr3_-_129375556 | 0.10 |
ENST00000510323.1
|
TMCC1
|
transmembrane and coiled-coil domain family 1 |
| chr2_-_47142884 | 0.10 |
ENST00000409105.1
ENST00000409973.1 ENST00000409913.1 ENST00000319466.4 |
MCFD2
|
multiple coagulation factor deficiency 2 |
| chr17_+_40440481 | 0.10 |
ENST00000590726.2
ENST00000452307.2 ENST00000444283.1 ENST00000588868.1 |
STAT5A
|
signal transducer and activator of transcription 5A |
| chr10_+_70715884 | 0.10 |
ENST00000354185.4
|
DDX21
|
DEAD (Asp-Glu-Ala-Asp) box helicase 21 |
| chr2_-_47143160 | 0.10 |
ENST00000409800.1
ENST00000409218.1 |
MCFD2
|
multiple coagulation factor deficiency 2 |
| chr8_+_38614754 | 0.10 |
ENST00000521642.1
|
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr6_+_151561506 | 0.10 |
ENST00000253332.1
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr2_+_187350973 | 0.10 |
ENST00000544130.1
|
ZC3H15
|
zinc finger CCCH-type containing 15 |
| chr5_+_34656331 | 0.10 |
ENST00000265109.3
|
RAI14
|
retinoic acid induced 14 |
| chr4_+_76649753 | 0.10 |
ENST00000603759.1
|
USO1
|
USO1 vesicle transport factor |
| chr7_+_111846741 | 0.10 |
ENST00000421043.1
ENST00000425229.1 ENST00000450657.1 |
ZNF277
|
zinc finger protein 277 |
| chr5_-_158636512 | 0.10 |
ENST00000424310.2
|
RNF145
|
ring finger protein 145 |
| chr17_-_38657849 | 0.10 |
ENST00000254051.6
|
TNS4
|
tensin 4 |
| chr11_+_18416103 | 0.10 |
ENST00000543445.1
ENST00000430553.2 ENST00000396222.2 ENST00000535451.1 |
LDHA
|
lactate dehydrogenase A |
| chr10_+_99609996 | 0.10 |
ENST00000370602.1
|
GOLGA7B
|
golgin A7 family, member B |
| chr7_+_116312411 | 0.10 |
ENST00000456159.1
ENST00000397752.3 ENST00000318493.6 |
MET
|
met proto-oncogene |
| chr3_+_66271410 | 0.09 |
ENST00000336733.6
|
SLC25A26
|
solute carrier family 25 (S-adenosylmethionine carrier), member 26 |
| chr11_+_18416133 | 0.09 |
ENST00000227157.4
ENST00000478970.2 ENST00000495052.1 |
LDHA
|
lactate dehydrogenase A |
| chr17_+_40985407 | 0.09 |
ENST00000586114.1
ENST00000590720.1 ENST00000585805.1 ENST00000541124.1 ENST00000441946.2 ENST00000591152.1 ENST00000589469.1 ENST00000293362.3 ENST00000592169.1 |
PSME3
|
proteasome (prosome, macropain) activator subunit 3 (PA28 gamma; Ki) |
| chr17_+_27052892 | 0.09 |
ENST00000579671.1
ENST00000579060.1 |
NEK8
|
NIMA-related kinase 8 |
| chr11_-_122932730 | 0.09 |
ENST00000532182.1
ENST00000524590.1 ENST00000528292.1 ENST00000533540.1 ENST00000525463.1 |
HSPA8
|
heat shock 70kDa protein 8 |
| chr9_-_19127474 | 0.09 |
ENST00000380465.3
ENST00000380464.3 ENST00000411567.1 ENST00000276914.2 |
PLIN2
|
perilipin 2 |
| chr4_-_76439483 | 0.09 |
ENST00000380840.2
ENST00000513257.1 ENST00000507014.1 |
RCHY1
|
ring finger and CHY zinc finger domain containing 1, E3 ubiquitin protein ligase |
| chr11_-_47870091 | 0.09 |
ENST00000526870.1
|
NUP160
|
nucleoporin 160kDa |
| chr7_+_26191809 | 0.09 |
ENST00000056233.3
|
NFE2L3
|
nuclear factor, erythroid 2-like 3 |
| chr13_-_95953589 | 0.09 |
ENST00000538287.1
ENST00000376887.4 ENST00000412704.1 ENST00000536256.1 ENST00000431522.1 |
ABCC4
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 4 |
| chr17_+_72428218 | 0.09 |
ENST00000392628.2
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr2_-_198364581 | 0.09 |
ENST00000428204.1
|
HSPD1
|
heat shock 60kDa protein 1 (chaperonin) |
| chr15_+_91446961 | 0.09 |
ENST00000559965.1
|
MAN2A2
|
mannosidase, alpha, class 2A, member 2 |
| chr6_+_151561085 | 0.08 |
ENST00000402676.2
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr11_+_133902547 | 0.08 |
ENST00000529070.1
|
RP11-713P17.3
|
RP11-713P17.3 |
| chr13_+_26828275 | 0.08 |
ENST00000381527.3
|
CDK8
|
cyclin-dependent kinase 8 |
| chr20_-_6103666 | 0.08 |
ENST00000536936.1
|
FERMT1
|
fermitin family member 1 |
| chr9_+_37079968 | 0.08 |
ENST00000588403.1
|
RP11-220I1.1
|
RP11-220I1.1 |
| chr6_-_41888843 | 0.08 |
ENST00000434077.1
ENST00000409312.1 |
MED20
|
mediator complex subunit 20 |
| chr4_-_103266626 | 0.08 |
ENST00000356736.4
|
SLC39A8
|
solute carrier family 39 (zinc transporter), member 8 |
| chr6_+_151646800 | 0.08 |
ENST00000354675.6
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr11_+_126081662 | 0.08 |
ENST00000528985.1
ENST00000529731.1 ENST00000360194.4 ENST00000530043.1 |
FAM118B
|
family with sequence similarity 118, member B |
| chr18_-_33047039 | 0.08 |
ENST00000591141.1
ENST00000586741.1 |
RP11-322E11.5
|
RP11-322E11.5 |
| chr10_-_7453445 | 0.08 |
ENST00000379713.3
ENST00000397167.1 ENST00000397160.3 |
SFMBT2
|
Scm-like with four mbt domains 2 |
| chr8_+_109455845 | 0.08 |
ENST00000220853.3
|
EMC2
|
ER membrane protein complex subunit 2 |
| chr15_-_49447771 | 0.08 |
ENST00000558843.1
ENST00000542928.1 ENST00000561248.1 |
COPS2
|
COP9 signalosome subunit 2 |
| chr17_+_30813576 | 0.08 |
ENST00000313401.3
|
CDK5R1
|
cyclin-dependent kinase 5, regulatory subunit 1 (p35) |
| chr15_-_52944231 | 0.08 |
ENST00000546305.2
|
FAM214A
|
family with sequence similarity 214, member A |
| chr11_-_45307817 | 0.08 |
ENST00000020926.3
|
SYT13
|
synaptotagmin XIII |
| chr7_-_45026159 | 0.08 |
ENST00000584327.1
ENST00000438705.3 |
SNHG15
|
small nucleolar RNA host gene 15 (non-protein coding) |
| chrX_-_128977875 | 0.08 |
ENST00000406492.2
|
ZDHHC9
|
zinc finger, DHHC-type containing 9 |
| chr11_+_69455855 | 0.08 |
ENST00000227507.2
ENST00000536559.1 |
CCND1
|
cyclin D1 |
| chr1_-_78149041 | 0.08 |
ENST00000414381.1
ENST00000370798.1 |
ZZZ3
|
zinc finger, ZZ-type containing 3 |
| chr19_+_55987998 | 0.08 |
ENST00000591164.1
|
ZNF628
|
zinc finger protein 628 |
| chr1_-_35325318 | 0.07 |
ENST00000423898.1
ENST00000456842.1 |
SMIM12
|
small integral membrane protein 12 |
| chr2_-_198364552 | 0.07 |
ENST00000439605.1
ENST00000418022.1 |
HSPD1
|
heat shock 60kDa protein 1 (chaperonin) |
| chr11_-_3013497 | 0.07 |
ENST00000448187.1
ENST00000532325.2 ENST00000399614.2 |
NAP1L4
|
nucleosome assembly protein 1-like 4 |
| chr12_+_64798095 | 0.07 |
ENST00000332707.5
|
XPOT
|
exportin, tRNA |
| chr2_+_10183651 | 0.07 |
ENST00000305883.1
|
KLF11
|
Kruppel-like factor 11 |
| chr11_+_72929402 | 0.07 |
ENST00000393596.2
|
P2RY2
|
purinergic receptor P2Y, G-protein coupled, 2 |
| chr6_+_149539053 | 0.07 |
ENST00000451095.1
|
RP1-111D6.3
|
RP1-111D6.3 |
| chr17_-_8059638 | 0.07 |
ENST00000584202.1
ENST00000354903.5 ENST00000577253.1 |
PER1
|
period circadian clock 1 |
| chr9_+_131903916 | 0.07 |
ENST00000419582.1
|
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr6_+_31553978 | 0.07 |
ENST00000376096.1
ENST00000376099.1 ENST00000376110.3 |
LST1
|
leukocyte specific transcript 1 |
| chr8_-_62627057 | 0.07 |
ENST00000519234.1
ENST00000379449.6 ENST00000379454.4 ENST00000518068.1 ENST00000517856.1 ENST00000356457.5 |
ASPH
|
aspartate beta-hydroxylase |
| chr9_+_37079888 | 0.07 |
ENST00000429493.1
ENST00000593237.1 ENST00000588557.1 ENST00000430809.1 ENST00000592157.1 |
RP11-220I1.1
|
RP11-220I1.1 |
| chr22_-_42310570 | 0.07 |
ENST00000457093.1
|
SHISA8
|
shisa family member 8 |
| chr3_+_149535022 | 0.07 |
ENST00000466795.1
|
RNF13
|
ring finger protein 13 |
| chr1_-_113498616 | 0.07 |
ENST00000433570.4
ENST00000538576.1 ENST00000458229.1 |
SLC16A1
|
solute carrier family 16 (monocarboxylate transporter), member 1 |
| chr5_-_133706695 | 0.07 |
ENST00000521755.1
ENST00000523054.1 ENST00000435240.2 ENST00000609654.1 ENST00000536186.1 ENST00000609383.1 |
CDKL3
|
cyclin-dependent kinase-like 3 |
| chr21_-_46492927 | 0.07 |
ENST00000599569.1
|
AP001579.1
|
Uncharacterized protein |
| chr3_+_112280857 | 0.07 |
ENST00000492406.1
ENST00000468642.1 |
SLC35A5
|
solute carrier family 35, member A5 |
| chr4_-_103266355 | 0.07 |
ENST00000424970.2
|
SLC39A8
|
solute carrier family 39 (zinc transporter), member 8 |
| chr19_+_46003056 | 0.07 |
ENST00000401593.1
ENST00000396736.2 |
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent, 1N (putative) |
| chr2_-_231084659 | 0.07 |
ENST00000258381.6
ENST00000358662.4 ENST00000455674.1 ENST00000392048.3 |
SP110
|
SP110 nuclear body protein |
| chr5_-_111312622 | 0.07 |
ENST00000395634.3
|
NREP
|
neuronal regeneration related protein |
| chr18_+_33877654 | 0.07 |
ENST00000257209.4
ENST00000445677.1 ENST00000590592.1 ENST00000359247.4 |
FHOD3
|
formin homology 2 domain containing 3 |
| chr15_-_45459704 | 0.06 |
ENST00000558039.1
|
CTD-2651B20.1
|
CTD-2651B20.1 |
| chr2_+_64069459 | 0.06 |
ENST00000445915.2
ENST00000475462.1 |
UGP2
|
UDP-glucose pyrophosphorylase 2 |
| chr9_+_131904233 | 0.06 |
ENST00000432651.1
ENST00000435132.1 |
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr15_+_74466012 | 0.06 |
ENST00000249842.3
|
ISLR
|
immunoglobulin superfamily containing leucine-rich repeat |
| chr16_+_66914264 | 0.06 |
ENST00000311765.2
ENST00000568869.1 ENST00000561704.1 ENST00000568398.1 ENST00000566776.1 |
PDP2
|
pyruvate dehyrogenase phosphatase catalytic subunit 2 |
| chr8_-_19540086 | 0.06 |
ENST00000332246.6
|
CSGALNACT1
|
chondroitin sulfate N-acetylgalactosaminyltransferase 1 |
| chr9_+_130159471 | 0.06 |
ENST00000419917.1
|
SLC2A8
|
solute carrier family 2 (facilitated glucose transporter), member 8 |
| chr12_+_49658855 | 0.06 |
ENST00000549183.1
|
TUBA1C
|
tubulin, alpha 1c |
| chr4_-_103266219 | 0.06 |
ENST00000394833.2
|
SLC39A8
|
solute carrier family 39 (zinc transporter), member 8 |
| chr17_-_76713100 | 0.06 |
ENST00000585509.1
|
CYTH1
|
cytohesin 1 |
| chr7_+_129007964 | 0.06 |
ENST00000460109.1
ENST00000474594.1 ENST00000446212.1 |
AHCYL2
|
adenosylhomocysteinase-like 2 |
| chr11_-_3013482 | 0.06 |
ENST00000529361.1
ENST00000528968.1 ENST00000534372.1 ENST00000531291.1 ENST00000526842.1 |
NAP1L4
|
nucleosome assembly protein 1-like 4 |
| chr12_+_6833323 | 0.06 |
ENST00000544725.1
|
COPS7A
|
COP9 signalosome subunit 7A |
| chr22_+_18593097 | 0.06 |
ENST00000426208.1
|
TUBA8
|
tubulin, alpha 8 |
| chr3_-_154042235 | 0.06 |
ENST00000308361.6
ENST00000496811.1 ENST00000544526.1 |
DHX36
|
DEAH (Asp-Glu-Ala-His) box polypeptide 36 |
| chr10_+_123970670 | 0.06 |
ENST00000496913.2
|
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr9_+_91933726 | 0.06 |
ENST00000534113.2
|
SECISBP2
|
SECIS binding protein 2 |
| chr3_-_142607740 | 0.06 |
ENST00000485766.1
|
PCOLCE2
|
procollagen C-endopeptidase enhancer 2 |
| chr9_-_88874519 | 0.06 |
ENST00000376001.3
ENST00000339137.3 |
C9orf153
|
chromosome 9 open reading frame 153 |
| chr21_-_16374688 | 0.06 |
ENST00000411932.1
|
NRIP1
|
nuclear receptor interacting protein 1 |
| chr2_+_227771404 | 0.06 |
ENST00000409053.1
|
RHBDD1
|
rhomboid domain containing 1 |
| chr17_+_60457914 | 0.06 |
ENST00000305286.3
ENST00000520404.1 ENST00000518576.1 |
EFCAB3
|
EF-hand calcium binding domain 3 |
| chr15_-_74495188 | 0.06 |
ENST00000563965.1
ENST00000395105.4 |
STRA6
|
stimulated by retinoic acid 6 |
| chr14_-_91884150 | 0.06 |
ENST00000553403.1
|
CCDC88C
|
coiled-coil domain containing 88C |
| chr18_+_44812072 | 0.06 |
ENST00000598649.1
ENST00000586905.2 |
CTD-2130O13.1
|
CTD-2130O13.1 |
| chr4_-_76439596 | 0.06 |
ENST00000451788.1
ENST00000512706.1 |
RCHY1
|
ring finger and CHY zinc finger domain containing 1, E3 ubiquitin protein ligase |
| chr11_-_47470682 | 0.06 |
ENST00000529341.1
ENST00000352508.3 |
RAPSN
|
receptor-associated protein of the synapse |
| chr2_+_17721920 | 0.06 |
ENST00000295156.4
|
VSNL1
|
visinin-like 1 |
| chr2_-_61389168 | 0.06 |
ENST00000607743.1
ENST00000605902.1 |
RP11-493E12.1
|
RP11-493E12.1 |
| chr3_+_188664988 | 0.06 |
ENST00000433971.1
|
TPRG1
|
tumor protein p63 regulated 1 |
| chr1_-_19229218 | 0.06 |
ENST00000432718.1
|
ALDH4A1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr11_-_122931881 | 0.06 |
ENST00000526110.1
ENST00000227378.3 |
HSPA8
|
heat shock 70kDa protein 8 |
| chr22_-_30695471 | 0.06 |
ENST00000434291.1
|
RP1-130H16.18
|
Uncharacterized protein |
| chr1_-_86174065 | 0.05 |
ENST00000370574.3
ENST00000431532.2 |
ZNHIT6
|
zinc finger, HIT-type containing 6 |
| chr6_+_142623758 | 0.05 |
ENST00000541199.1
ENST00000435011.2 |
GPR126
|
G protein-coupled receptor 126 |
| chr3_+_57741957 | 0.05 |
ENST00000295951.3
|
SLMAP
|
sarcolemma associated protein |
| chr12_-_76477707 | 0.05 |
ENST00000551992.1
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr5_-_93447333 | 0.05 |
ENST00000395965.3
ENST00000505869.1 ENST00000509163.1 |
FAM172A
|
family with sequence similarity 172, member A |
| chr16_+_56703703 | 0.05 |
ENST00000332374.4
|
MT1H
|
metallothionein 1H |
| chr22_+_18560743 | 0.05 |
ENST00000399744.3
|
PEX26
|
peroxisomal biogenesis factor 26 |
| chr19_-_1237990 | 0.05 |
ENST00000382477.2
ENST00000215376.6 ENST00000590083.1 |
C19orf26
|
chromosome 19 open reading frame 26 |
| chr12_-_123634449 | 0.05 |
ENST00000542210.1
|
PITPNM2
|
phosphatidylinositol transfer protein, membrane-associated 2 |
| chr7_+_106809406 | 0.05 |
ENST00000468410.1
ENST00000478930.1 ENST00000464009.1 ENST00000222574.4 |
HBP1
|
HMG-box transcription factor 1 |
| chr11_-_71814276 | 0.05 |
ENST00000538404.1
ENST00000535107.1 ENST00000545249.1 |
LAMTOR1
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 1 |
| chr1_+_166808692 | 0.05 |
ENST00000367876.4
|
POGK
|
pogo transposable element with KRAB domain |
| chr21_-_45079341 | 0.05 |
ENST00000443485.1
ENST00000291560.2 |
HSF2BP
|
heat shock transcription factor 2 binding protein |
| chr15_-_49447835 | 0.05 |
ENST00000388901.5
ENST00000299259.6 |
COPS2
|
COP9 signalosome subunit 2 |
| chr2_-_55276320 | 0.05 |
ENST00000357376.3
|
RTN4
|
reticulon 4 |
| chr5_+_70883154 | 0.05 |
ENST00000509358.2
|
MCCC2
|
methylcrotonoyl-CoA carboxylase 2 (beta) |
| chr2_-_231084820 | 0.05 |
ENST00000258382.5
ENST00000338556.3 |
SP110
|
SP110 nuclear body protein |
| chr19_+_48949087 | 0.05 |
ENST00000598711.1
|
GRWD1
|
glutamate-rich WD repeat containing 1 |
| chr8_-_130951940 | 0.05 |
ENST00000522250.1
ENST00000522941.1 ENST00000522746.1 ENST00000520204.1 ENST00000519070.1 ENST00000520254.1 ENST00000519824.2 ENST00000519540.1 |
FAM49B
|
family with sequence similarity 49, member B |
| chr11_-_126081532 | 0.05 |
ENST00000533628.1
ENST00000298317.4 ENST00000532674.1 |
RPUSD4
|
RNA pseudouridylate synthase domain containing 4 |
| chr4_+_57333756 | 0.05 |
ENST00000510663.1
ENST00000504757.1 |
SRP72
|
signal recognition particle 72kDa |
| chr20_+_42839600 | 0.05 |
ENST00000439943.1
ENST00000437730.1 |
OSER1-AS1
|
OSER1 antisense RNA 1 (head to head) |
| chr16_-_47493041 | 0.05 |
ENST00000565940.2
|
ITFG1
|
integrin alpha FG-GAP repeat containing 1 |
| chr2_-_216257849 | 0.05 |
ENST00000456923.1
|
FN1
|
fibronectin 1 |
| chr21_-_35987438 | 0.05 |
ENST00000313806.4
|
RCAN1
|
regulator of calcineurin 1 |
| chr5_-_115177247 | 0.05 |
ENST00000500945.2
|
ATG12
|
autophagy related 12 |
| chr1_-_111991908 | 0.05 |
ENST00000235090.5
|
WDR77
|
WD repeat domain 77 |
| chr2_+_118572226 | 0.05 |
ENST00000263239.2
|
DDX18
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 18 |
| chrX_+_54556633 | 0.05 |
ENST00000336470.4
ENST00000360845.2 |
GNL3L
|
guanine nucleotide binding protein-like 3 (nucleolar)-like |
| chr6_-_90529418 | 0.05 |
ENST00000439638.1
ENST00000369393.3 ENST00000428876.1 |
MDN1
|
MDN1, midasin homolog (yeast) |
| chr2_-_158295915 | 0.05 |
ENST00000418920.1
|
CYTIP
|
cytohesin 1 interacting protein |
| chr11_-_74022658 | 0.05 |
ENST00000427714.2
ENST00000331597.4 |
P4HA3
|
prolyl 4-hydroxylase, alpha polypeptide III |
| chr16_-_2162831 | 0.05 |
ENST00000483024.1
|
PKD1
|
polycystic kidney disease 1 (autosomal dominant) |
| chr19_+_19516561 | 0.05 |
ENST00000457895.2
|
GATAD2A
|
GATA zinc finger domain containing 2A |
| chr7_+_30791743 | 0.05 |
ENST00000013222.5
ENST00000409539.1 |
INMT
|
indolethylamine N-methyltransferase |
| chr9_+_131904295 | 0.05 |
ENST00000434095.1
|
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr6_-_43496605 | 0.05 |
ENST00000455285.2
|
XPO5
|
exportin 5 |
| chr1_-_154193009 | 0.05 |
ENST00000368518.1
ENST00000368519.1 ENST00000368521.5 |
C1orf43
|
chromosome 1 open reading frame 43 |
| chr12_+_57623869 | 0.05 |
ENST00000414700.3
ENST00000557703.1 |
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr12_-_31478428 | 0.05 |
ENST00000543615.1
|
FAM60A
|
family with sequence similarity 60, member A |
| chr13_+_98628886 | 0.05 |
ENST00000490680.1
ENST00000539640.1 ENST00000403772.3 |
IPO5
|
importin 5 |
| chr15_-_82555000 | 0.05 |
ENST00000557844.1
ENST00000359445.3 ENST00000268206.7 |
EFTUD1
|
elongation factor Tu GTP binding domain containing 1 |
| chr5_+_138609441 | 0.05 |
ENST00000509990.1
ENST00000506147.1 ENST00000512107.1 |
MATR3
|
matrin 3 |
| chr11_-_94226964 | 0.05 |
ENST00000538923.1
ENST00000540013.1 ENST00000407439.3 ENST00000393241.4 |
MRE11A
|
MRE11 meiotic recombination 11 homolog A (S. cerevisiae) |
| chrX_+_23685653 | 0.05 |
ENST00000379331.3
|
PRDX4
|
peroxiredoxin 4 |
| chr4_-_4291761 | 0.05 |
ENST00000513174.1
|
LYAR
|
Ly1 antibody reactive |
| chr17_+_72427477 | 0.05 |
ENST00000342648.5
ENST00000481232.1 |
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr1_+_100436065 | 0.05 |
ENST00000370153.1
|
SLC35A3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3 |
| chr9_-_98268883 | 0.05 |
ENST00000551630.1
ENST00000548420.1 |
PTCH1
|
patched 1 |
| chr18_+_9136758 | 0.05 |
ENST00000383440.2
ENST00000262126.4 ENST00000577992.1 |
ANKRD12
|
ankyrin repeat domain 12 |
| chr2_+_201171372 | 0.05 |
ENST00000409140.3
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr2_-_231084617 | 0.05 |
ENST00000409815.2
|
SP110
|
SP110 nuclear body protein |
| chr9_-_34637806 | 0.04 |
ENST00000477726.1
|
SIGMAR1
|
sigma non-opioid intracellular receptor 1 |
| chr11_-_107436443 | 0.04 |
ENST00000429370.1
ENST00000417449.2 ENST00000428149.2 |
ALKBH8
|
alkB, alkylation repair homolog 8 (E. coli) |
| chr11_-_64511789 | 0.04 |
ENST00000419843.1
ENST00000394430.1 |
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr5_+_110427983 | 0.04 |
ENST00000513710.2
ENST00000505303.1 |
WDR36
|
WD repeat domain 36 |
| chr8_+_104383759 | 0.04 |
ENST00000415886.2
|
CTHRC1
|
collagen triple helix repeat containing 1 |
| chr6_-_47277634 | 0.04 |
ENST00000296861.2
|
TNFRSF21
|
tumor necrosis factor receptor superfamily, member 21 |
| chr12_+_52668394 | 0.04 |
ENST00000423955.2
|
KRT86
|
keratin 86 |
| chr5_+_96077888 | 0.04 |
ENST00000509259.1
ENST00000503828.1 |
CAST
|
calpastatin |
Network of associatons between targets according to the STRING database.
First level regulatory network of CLOCK
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0002368 | B cell cytokine production(GO:0002368) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.3 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.0 | 0.2 | GO:0072425 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.0 | 0.1 | GO:0046901 | tetrahydrofolylpolyglutamate biosynthetic process(GO:0046901) |
| 0.0 | 0.1 | GO:1902904 | negative regulation of fibril organization(GO:1902904) chaperone-mediated autophagy translocation complex disassembly(GO:1904764) |
| 0.0 | 0.1 | GO:0021718 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.0 | 0.1 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.0 | 0.1 | GO:0015728 | mevalonate transport(GO:0015728) behavioral response to nutrient(GO:0051780) |
| 0.0 | 0.0 | GO:2001180 | negative regulation of interleukin-10 secretion(GO:2001180) |
| 0.0 | 0.1 | GO:0006311 | meiotic gene conversion(GO:0006311) |
| 0.0 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.0 | 0.2 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.3 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.1 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.2 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.1 | GO:1904235 | regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904235) positive regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904237) |
| 0.0 | 0.1 | GO:2001201 | regulation of transforming growth factor-beta secretion(GO:2001201) |
| 0.0 | 0.0 | GO:0035281 | pre-miRNA export from nucleus(GO:0035281) |
| 0.0 | 0.0 | GO:0007057 | spindle assembly involved in female meiosis I(GO:0007057) |
| 0.0 | 0.0 | GO:0006258 | UDP-glucose catabolic process(GO:0006258) |
| 0.0 | 0.1 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) |
| 0.0 | 0.2 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.0 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.0 | 0.0 | GO:0000961 | negative regulation of mitochondrial RNA catabolic process(GO:0000961) |
| 0.0 | 0.1 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.2 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.1 | GO:2000909 | regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.0 | 0.0 | GO:0003220 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) |
| 0.0 | 0.0 | GO:0072287 | metanephric distal tubule morphogenesis(GO:0072287) |
| 0.0 | 0.1 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.0 | 0.1 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.0 | 0.2 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.0 | GO:1900073 | regulation of neuromuscular synaptic transmission(GO:1900073) positive regulation of neuromuscular synaptic transmission(GO:1900075) |
| 0.0 | 0.1 | GO:0043697 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) negative regulation of monocyte differentiation(GO:0045656) |
| 0.0 | 0.0 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.0 | 0.0 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.0 | 0.3 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.1 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.1 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.0 | GO:0042565 | RNA nuclear export complex(GO:0042565) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.1 | GO:0097362 | MCM8-MCM9 complex(GO:0097362) |
| 0.0 | 0.1 | GO:0031515 | tRNA (m1A) methyltransferase complex(GO:0031515) |
| 0.0 | 0.2 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.0 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.1 | 0.2 | GO:0043849 | Ras palmitoyltransferase activity(GO:0043849) |
| 0.0 | 0.1 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.0 | 0.1 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.3 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.0 | 0.1 | GO:0008841 | tetrahydrofolylpolyglutamate synthase activity(GO:0004326) dihydrofolate synthase activity(GO:0008841) |
| 0.0 | 0.2 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.0 | 0.2 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0015130 | mevalonate transmembrane transporter activity(GO:0015130) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.1 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.0 | 0.2 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.2 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.0 | 0.1 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.0 | GO:0090631 | pre-miRNA transporter activity(GO:0090631) |
| 0.0 | 0.1 | GO:0004572 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity(GO:0004572) |
| 0.0 | 0.1 | GO:0033765 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 0.3 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.1 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.0 | 0.0 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.0 | 0.0 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.0 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.4 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.1 | REACTOME G1 PHASE | Genes involved in G1 Phase |