Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
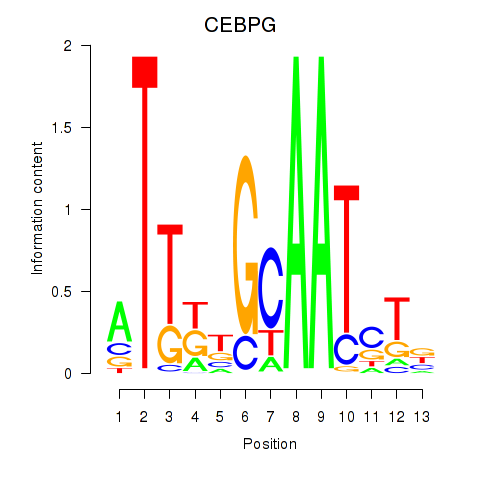
Results for CEBPG
Z-value: 1.45
Transcription factors associated with CEBPG
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CEBPG
|
ENSG00000153879.4 | CCAAT enhancer binding protein gamma |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CEBPG | hg19_v2_chr19_+_33865218_33865254 | -0.89 | 1.1e-01 | Click! |
Activity profile of CEBPG motif
Sorted Z-values of CEBPG motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_102668879 | 2.96 |
ENST00000315274.6
|
MMP1
|
matrix metallopeptidase 1 (interstitial collagenase) |
| chr19_+_39786962 | 2.93 |
ENST00000333625.2
|
IFNL1
|
interferon, lambda 1 |
| chr1_+_158979680 | 1.78 |
ENST00000368131.4
ENST00000340979.6 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr11_-_33913708 | 1.74 |
ENST00000257818.2
|
LMO2
|
LIM domain only 2 (rhombotin-like 1) |
| chr2_-_191885686 | 1.73 |
ENST00000432058.1
|
STAT1
|
signal transducer and activator of transcription 1, 91kDa |
| chr1_+_158979686 | 1.72 |
ENST00000368132.3
ENST00000295809.7 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr1_+_158979792 | 1.71 |
ENST00000359709.3
ENST00000430894.2 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr19_-_1155118 | 1.69 |
ENST00000590998.1
|
SBNO2
|
strawberry notch homolog 2 (Drosophila) |
| chr7_+_22766766 | 1.67 |
ENST00000426291.1
ENST00000401651.1 ENST00000258743.5 ENST00000420258.2 ENST00000407492.1 ENST00000401630.3 ENST00000406575.1 |
IL6
|
interleukin 6 (interferon, beta 2) |
| chr6_+_31895480 | 1.45 |
ENST00000418949.2
ENST00000383177.3 ENST00000477310.1 |
C2
CFB
|
complement component 2 Complement factor B; Uncharacterized protein; cDNA FLJ55673, highly similar to Complement factor B |
| chr6_+_31895467 | 1.44 |
ENST00000556679.1
ENST00000456570.1 |
CFB
CFB
|
complement factor B Complement factor B; Uncharacterized protein; cDNA FLJ55673, highly similar to Complement factor B |
| chr11_-_104972158 | 1.44 |
ENST00000598974.1
ENST00000593315.1 ENST00000594519.1 ENST00000415981.2 ENST00000525374.1 ENST00000375707.1 |
CASP1
CARD16
CARD17
|
caspase 1, apoptosis-related cysteine peptidase caspase recruitment domain family, member 16 caspase recruitment domain family, member 17 |
| chr11_+_18287801 | 1.40 |
ENST00000532858.1
ENST00000405158.2 |
SAA1
|
serum amyloid A1 |
| chr11_-_18270182 | 1.39 |
ENST00000528349.1
ENST00000526900.1 ENST00000529528.1 ENST00000414546.2 ENST00000256733.4 |
SAA2
|
serum amyloid A2 |
| chr11_-_104916034 | 1.38 |
ENST00000528513.1
ENST00000375706.2 ENST00000375704.3 |
CARD16
|
caspase recruitment domain family, member 16 |
| chr11_+_18287721 | 1.24 |
ENST00000356524.4
|
SAA1
|
serum amyloid A1 |
| chr19_+_6887571 | 1.20 |
ENST00000250572.8
ENST00000381407.5 ENST00000312053.4 ENST00000450315.3 ENST00000381404.4 |
EMR1
|
egf-like module containing, mucin-like, hormone receptor-like 1 |
| chr2_-_152146385 | 1.14 |
ENST00000414946.1
ENST00000243346.5 |
NMI
|
N-myc (and STAT) interactor |
| chr11_-_18258342 | 1.03 |
ENST00000278222.4
|
SAA4
|
serum amyloid A4, constitutive |
| chr5_-_121413974 | 0.92 |
ENST00000231004.4
|
LOX
|
lysyl oxidase |
| chr11_-_124806297 | 0.75 |
ENST00000298251.4
|
HEPACAM
|
hepatic and glial cell adhesion molecule |
| chr19_-_6720686 | 0.71 |
ENST00000245907.6
|
C3
|
complement component 3 |
| chr12_-_69080590 | 0.70 |
ENST00000433116.2
ENST00000500695.2 |
RP11-637A17.2
|
RP11-637A17.2 |
| chr6_+_31895254 | 0.70 |
ENST00000299367.5
ENST00000442278.2 |
C2
|
complement component 2 |
| chr2_+_65663812 | 0.68 |
ENST00000606978.1
ENST00000377977.3 ENST00000536804.1 |
AC074391.1
|
AC074391.1 |
| chr9_+_130911723 | 0.63 |
ENST00000277480.2
ENST00000373013.2 ENST00000540948.1 |
LCN2
|
lipocalin 2 |
| chr6_-_133055896 | 0.62 |
ENST00000367927.5
ENST00000425515.2 ENST00000207771.3 ENST00000392393.3 ENST00000450865.2 ENST00000392394.2 |
VNN3
|
vanin 3 |
| chr21_+_33671160 | 0.61 |
ENST00000303645.5
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr9_+_130911770 | 0.61 |
ENST00000372998.1
|
LCN2
|
lipocalin 2 |
| chr21_+_34775181 | 0.59 |
ENST00000290219.6
|
IFNGR2
|
interferon gamma receptor 2 (interferon gamma transducer 1) |
| chr1_-_47655686 | 0.58 |
ENST00000294338.2
|
PDZK1IP1
|
PDZK1 interacting protein 1 |
| chr21_+_33671264 | 0.57 |
ENST00000339944.4
|
MRAP
|
melanocortin 2 receptor accessory protein |
| chr17_-_39769005 | 0.56 |
ENST00000301653.4
ENST00000593067.1 |
KRT16
|
keratin 16 |
| chr12_-_96390108 | 0.56 |
ENST00000538703.1
ENST00000261208.3 |
HAL
|
histidine ammonia-lyase |
| chr6_-_133055815 | 0.53 |
ENST00000509351.1
ENST00000417437.2 ENST00000414302.2 ENST00000423615.2 ENST00000427187.2 ENST00000275223.3 ENST00000519686.2 |
VNN3
|
vanin 3 |
| chr5_+_49962772 | 0.52 |
ENST00000281631.5
ENST00000513738.1 ENST00000503665.1 ENST00000514067.2 ENST00000503046.1 |
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr2_-_32390801 | 0.47 |
ENST00000608489.1
|
RP11-563N4.1
|
RP11-563N4.1 |
| chr12_-_54758251 | 0.47 |
ENST00000267015.3
ENST00000551809.1 |
GPR84
|
G protein-coupled receptor 84 |
| chrX_+_46937745 | 0.46 |
ENST00000397180.1
ENST00000457380.1 ENST00000352078.4 |
RGN
|
regucalcin |
| chrX_-_73512177 | 0.46 |
ENST00000603672.1
ENST00000418855.1 |
FTX
|
FTX transcript, XIST regulator (non-protein coding) |
| chr5_+_125758813 | 0.46 |
ENST00000285689.3
ENST00000515200.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr5_+_125758865 | 0.43 |
ENST00000542322.1
ENST00000544396.1 |
GRAMD3
|
GRAM domain containing 3 |
| chr9_+_110045418 | 0.43 |
ENST00000419616.1
|
RAD23B
|
RAD23 homolog B (S. cerevisiae) |
| chr7_+_137761199 | 0.43 |
ENST00000411726.2
|
AKR1D1
|
aldo-keto reductase family 1, member D1 |
| chr12_-_8693469 | 0.42 |
ENST00000545274.1
ENST00000446457.2 |
CLEC4E
|
C-type lectin domain family 4, member E |
| chr21_+_43619796 | 0.40 |
ENST00000398457.2
|
ABCG1
|
ATP-binding cassette, sub-family G (WHITE), member 1 |
| chr14_+_62462541 | 0.38 |
ENST00000430451.2
|
SYT16
|
synaptotagmin XVI |
| chr17_+_812872 | 0.36 |
ENST00000576252.1
|
RP11-676J12.7
|
Uncharacterized protein |
| chr2_-_113594279 | 0.34 |
ENST00000416750.1
ENST00000418817.1 ENST00000263341.2 |
IL1B
|
interleukin 1, beta |
| chr9_+_130860810 | 0.33 |
ENST00000433501.1
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25 |
| chr1_+_52682052 | 0.32 |
ENST00000371591.1
|
ZFYVE9
|
zinc finger, FYVE domain containing 9 |
| chr16_+_57220049 | 0.32 |
ENST00000562439.1
|
RSPRY1
|
ring finger and SPRY domain containing 1 |
| chr19_-_39805976 | 0.32 |
ENST00000248668.4
|
LRFN1
|
leucine rich repeat and fibronectin type III domain containing 1 |
| chr12_+_53835425 | 0.30 |
ENST00000549924.1
|
PRR13
|
proline rich 13 |
| chr16_-_57219721 | 0.29 |
ENST00000562406.1
ENST00000568671.1 ENST00000567044.1 |
FAM192A
|
family with sequence similarity 192, member A |
| chr17_+_4336955 | 0.29 |
ENST00000355530.2
|
SPNS3
|
spinster homolog 3 (Drosophila) |
| chr12_-_8693539 | 0.29 |
ENST00000299663.3
|
CLEC4E
|
C-type lectin domain family 4, member E |
| chr17_-_76124711 | 0.28 |
ENST00000306591.7
ENST00000590602.1 |
TMC6
|
transmembrane channel-like 6 |
| chr11_+_76493294 | 0.28 |
ENST00000533752.1
|
TSKU
|
tsukushi, small leucine rich proteoglycan |
| chr9_+_130860583 | 0.28 |
ENST00000373064.5
|
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25 |
| chr7_-_8276508 | 0.28 |
ENST00000401396.1
ENST00000317367.5 |
ICA1
|
islet cell autoantigen 1, 69kDa |
| chr17_+_57287228 | 0.28 |
ENST00000578922.1
ENST00000300917.5 |
SMG8
|
SMG8 nonsense mediated mRNA decay factor |
| chr12_-_96390063 | 0.28 |
ENST00000541929.1
|
HAL
|
histidine ammonia-lyase |
| chr13_+_30002741 | 0.26 |
ENST00000380808.2
|
MTUS2
|
microtubule associated tumor suppressor candidate 2 |
| chr22_-_50523760 | 0.26 |
ENST00000395876.2
|
MLC1
|
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr3_-_157221128 | 0.26 |
ENST00000392833.2
ENST00000362010.2 |
VEPH1
|
ventricular zone expressed PH domain-containing 1 |
| chr17_+_4699439 | 0.25 |
ENST00000270586.3
|
PSMB6
|
proteasome (prosome, macropain) subunit, beta type, 6 |
| chr5_+_125759140 | 0.25 |
ENST00000543198.1
|
GRAMD3
|
GRAM domain containing 3 |
| chr12_+_57522439 | 0.25 |
ENST00000338962.4
|
LRP1
|
low density lipoprotein receptor-related protein 1 |
| chr18_-_29340827 | 0.24 |
ENST00000269205.5
|
SLC25A52
|
solute carrier family 25, member 52 |
| chr22_-_31364187 | 0.24 |
ENST00000215862.4
ENST00000397641.3 |
MORC2
|
MORC family CW-type zinc finger 2 |
| chr11_-_26593649 | 0.23 |
ENST00000455601.2
|
MUC15
|
mucin 15, cell surface associated |
| chr6_+_30749649 | 0.23 |
ENST00000422944.1
|
HCG20
|
HLA complex group 20 (non-protein coding) |
| chr17_+_4337199 | 0.23 |
ENST00000333476.2
|
SPNS3
|
spinster homolog 3 (Drosophila) |
| chr5_-_94417562 | 0.22 |
ENST00000505465.1
|
MCTP1
|
multiple C2 domains, transmembrane 1 |
| chr6_+_36238237 | 0.22 |
ENST00000457797.1
ENST00000394571.2 |
PNPLA1
|
patatin-like phospholipase domain containing 1 |
| chr7_-_107968999 | 0.22 |
ENST00000456431.1
|
NRCAM
|
neuronal cell adhesion molecule |
| chr20_+_361261 | 0.22 |
ENST00000217233.3
|
TRIB3
|
tribbles pseudokinase 3 |
| chr1_+_172389821 | 0.21 |
ENST00000367727.4
|
C1orf105
|
chromosome 1 open reading frame 105 |
| chr3_-_42306248 | 0.21 |
ENST00000334681.5
|
CCK
|
cholecystokinin |
| chr5_+_49963239 | 0.21 |
ENST00000505554.1
|
PARP8
|
poly (ADP-ribose) polymerase family, member 8 |
| chr14_-_70263979 | 0.20 |
ENST00000216540.4
|
SLC10A1
|
solute carrier family 10 (sodium/bile acid cotransporter), member 1 |
| chr1_-_204183071 | 0.20 |
ENST00000308302.3
|
GOLT1A
|
golgi transport 1A |
| chr11_+_60048129 | 0.19 |
ENST00000355131.3
|
MS4A4A
|
membrane-spanning 4-domains, subfamily A, member 4A |
| chr4_+_184020398 | 0.19 |
ENST00000403733.3
ENST00000378925.3 |
WWC2
|
WW and C2 domain containing 2 |
| chr7_-_107968921 | 0.19 |
ENST00000442580.1
|
NRCAM
|
neuronal cell adhesion molecule |
| chr7_-_138386097 | 0.19 |
ENST00000421622.1
|
SVOPL
|
SVOP-like |
| chr11_-_5248294 | 0.19 |
ENST00000335295.4
|
HBB
|
hemoglobin, beta |
| chr11_+_60048053 | 0.19 |
ENST00000337908.4
|
MS4A4A
|
membrane-spanning 4-domains, subfamily A, member 4A |
| chr14_+_96722539 | 0.18 |
ENST00000553356.1
|
BDKRB1
|
bradykinin receptor B1 |
| chr17_+_2699697 | 0.18 |
ENST00000254695.8
ENST00000366401.4 ENST00000542807.1 |
RAP1GAP2
|
RAP1 GTPase activating protein 2 |
| chr18_+_72167096 | 0.18 |
ENST00000324301.8
|
CNDP2
|
CNDP dipeptidase 2 (metallopeptidase M20 family) |
| chr19_-_6670128 | 0.16 |
ENST00000245912.3
|
TNFSF14
|
tumor necrosis factor (ligand) superfamily, member 14 |
| chr1_-_179112173 | 0.16 |
ENST00000408940.3
ENST00000504405.1 |
ABL2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr16_-_57219966 | 0.16 |
ENST00000565760.1
ENST00000309137.8 ENST00000570184.1 ENST00000562324.1 |
FAM192A
|
family with sequence similarity 192, member A |
| chr12_-_10605929 | 0.16 |
ENST00000347831.5
ENST00000359151.3 |
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1 |
| chr6_-_30080863 | 0.16 |
ENST00000540829.1
|
TRIM31
|
tripartite motif containing 31 |
| chr6_+_31895287 | 0.15 |
ENST00000447952.2
|
C2
|
complement component 2 |
| chr1_+_111770232 | 0.15 |
ENST00000369744.2
|
CHI3L2
|
chitinase 3-like 2 |
| chr7_-_14026063 | 0.15 |
ENST00000443608.1
ENST00000438956.1 |
ETV1
|
ets variant 1 |
| chr9_+_136399929 | 0.15 |
ENST00000393060.1
|
ADAMTSL2
|
ADAMTS-like 2 |
| chr19_-_49496557 | 0.15 |
ENST00000323798.3
ENST00000541188.1 ENST00000544287.1 ENST00000540532.1 ENST00000263276.6 |
GYS1
|
glycogen synthase 1 (muscle) |
| chr7_-_83824169 | 0.15 |
ENST00000265362.4
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr2_+_226265364 | 0.14 |
ENST00000272907.6
|
NYAP2
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 2 |
| chr21_-_37852359 | 0.14 |
ENST00000399137.1
ENST00000399135.1 |
CLDN14
|
claudin 14 |
| chr3_+_40498783 | 0.14 |
ENST00000338970.6
ENST00000396203.2 ENST00000416518.1 |
RPL14
|
ribosomal protein L14 |
| chr19_+_37808831 | 0.14 |
ENST00000589801.1
|
HKR1
|
HKR1, GLI-Kruppel zinc finger family member |
| chr1_-_1711508 | 0.13 |
ENST00000378625.1
|
NADK
|
NAD kinase |
| chr1_-_179112189 | 0.13 |
ENST00000512653.1
ENST00000344730.3 |
ABL2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr15_+_67813406 | 0.13 |
ENST00000342683.4
|
C15orf61
|
chromosome 15 open reading frame 61 |
| chr19_-_36822595 | 0.13 |
ENST00000585356.1
ENST00000438368.2 ENST00000590622.1 |
LINC00665
|
long intergenic non-protein coding RNA 665 |
| chr1_-_17676070 | 0.13 |
ENST00000602074.1
|
AC004824.2
|
Uncharacterized protein |
| chr2_+_120303717 | 0.13 |
ENST00000594371.1
|
PCDP1
|
Primary ciliary dyskinesia protein 1 |
| chr10_+_26986582 | 0.12 |
ENST00000376215.5
ENST00000376203.5 |
PDSS1
|
prenyl (decaprenyl) diphosphate synthase, subunit 1 |
| chr11_-_72504681 | 0.11 |
ENST00000538536.1
ENST00000543304.1 ENST00000540587.1 ENST00000334805.6 |
STARD10
|
StAR-related lipid transfer (START) domain containing 10 |
| chr6_-_30080876 | 0.11 |
ENST00000376734.3
|
TRIM31
|
tripartite motif containing 31 |
| chr16_-_57219926 | 0.11 |
ENST00000566584.1
ENST00000566481.1 ENST00000566077.1 ENST00000564108.1 ENST00000565458.1 ENST00000566681.1 ENST00000567439.1 |
FAM192A
|
family with sequence similarity 192, member A |
| chr9_-_86323118 | 0.10 |
ENST00000376395.4
|
UBQLN1
|
ubiquilin 1 |
| chr10_+_18629628 | 0.10 |
ENST00000377329.4
|
CACNB2
|
calcium channel, voltage-dependent, beta 2 subunit |
| chr20_+_361890 | 0.10 |
ENST00000449710.1
ENST00000422053.2 |
TRIB3
|
tribbles pseudokinase 3 |
| chr12_-_95510743 | 0.10 |
ENST00000551521.1
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr3_+_157154578 | 0.09 |
ENST00000295927.3
|
PTX3
|
pentraxin 3, long |
| chr20_-_60294804 | 0.09 |
ENST00000317652.1
|
RP11-429E11.3
|
Uncharacterized protein |
| chr2_-_175711978 | 0.09 |
ENST00000409089.2
|
CHN1
|
chimerin 1 |
| chr2_-_112614424 | 0.09 |
ENST00000427997.1
|
ANAPC1
|
anaphase promoting complex subunit 1 |
| chr7_-_144435985 | 0.08 |
ENST00000549981.1
|
TPK1
|
thiamin pyrophosphokinase 1 |
| chr17_-_29645836 | 0.08 |
ENST00000578584.1
|
CTD-2370N5.3
|
CTD-2370N5.3 |
| chr9_+_117092149 | 0.08 |
ENST00000431067.2
ENST00000412657.1 |
ORM2
|
orosomucoid 2 |
| chr12_-_53074182 | 0.07 |
ENST00000252244.3
|
KRT1
|
keratin 1 |
| chr20_+_54987168 | 0.07 |
ENST00000360314.3
|
CASS4
|
Cas scaffolding protein family member 4 |
| chr8_+_38586068 | 0.07 |
ENST00000443286.2
ENST00000520340.1 ENST00000518415.1 |
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr5_-_179050066 | 0.06 |
ENST00000329433.6
ENST00000510411.1 |
HNRNPH1
|
heterogeneous nuclear ribonucleoprotein H1 (H) |
| chr14_+_45366518 | 0.06 |
ENST00000557112.1
|
C14orf28
|
chromosome 14 open reading frame 28 |
| chr9_+_115913222 | 0.06 |
ENST00000259392.3
|
SLC31A2
|
solute carrier family 31 (copper transporter), member 2 |
| chr3_+_73110810 | 0.05 |
ENST00000533473.1
|
EBLN2
|
endogenous Bornavirus-like nucleoprotein 2 |
| chr1_-_153321301 | 0.05 |
ENST00000368739.3
|
PGLYRP4
|
peptidoglycan recognition protein 4 |
| chr14_+_39703112 | 0.04 |
ENST00000555143.1
ENST00000280082.3 |
MIA2
|
melanoma inhibitory activity 2 |
| chr12_+_53835383 | 0.04 |
ENST00000429243.2
|
PRR13
|
proline rich 13 |
| chr9_+_110045537 | 0.04 |
ENST00000358015.3
|
RAD23B
|
RAD23 homolog B (S. cerevisiae) |
| chr3_-_49726104 | 0.04 |
ENST00000383728.3
ENST00000545762.1 |
MST1
|
macrophage stimulating 1 (hepatocyte growth factor-like) |
| chr12_-_13248598 | 0.04 |
ENST00000337630.6
ENST00000545699.1 |
GSG1
|
germ cell associated 1 |
| chr4_-_111119804 | 0.04 |
ENST00000394607.3
ENST00000302274.3 |
ELOVL6
|
ELOVL fatty acid elongase 6 |
| chr4_-_10023095 | 0.03 |
ENST00000264784.3
|
SLC2A9
|
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chrX_-_73834449 | 0.03 |
ENST00000332687.6
ENST00000349225.2 |
RLIM
|
ring finger protein, LIM domain interacting |
| chr5_+_36152091 | 0.03 |
ENST00000274254.5
|
SKP2
|
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr6_-_27799305 | 0.03 |
ENST00000357549.2
|
HIST1H4K
|
histone cluster 1, H4k |
| chr5_+_36152179 | 0.02 |
ENST00000508514.1
ENST00000513151.1 ENST00000546211.1 |
SKP2
|
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr19_+_49496782 | 0.01 |
ENST00000601968.1
ENST00000596837.1 |
RUVBL2
|
RuvB-like AAA ATPase 2 |
| chr3_+_186435137 | 0.01 |
ENST00000447445.1
|
KNG1
|
kininogen 1 |
| chr1_+_158901329 | 0.01 |
ENST00000368140.1
ENST00000368138.3 ENST00000392254.2 ENST00000392252.3 ENST00000368135.4 |
PYHIN1
|
pyrin and HIN domain family, member 1 |
| chr19_-_54327542 | 0.01 |
ENST00000391775.3
ENST00000324134.6 ENST00000535162.1 ENST00000351894.4 ENST00000354278.3 ENST00000391773.1 ENST00000345770.5 ENST00000391772.1 |
NLRP12
|
NLR family, pyrin domain containing 12 |
| chr17_+_7477040 | 0.00 |
ENST00000581384.1
ENST00000577929.1 |
EIF4A1
|
eukaryotic translation initiation factor 4A1 |
| chr7_-_14026123 | 0.00 |
ENST00000420159.2
ENST00000399357.3 ENST00000403527.1 |
ETV1
|
ets variant 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of CEBPG
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.9 | GO:0032696 | negative regulation of interleukin-13 production(GO:0032696) |
| 0.6 | 1.7 | GO:2000366 | regulation of STAT protein import into nucleus(GO:2000364) positive regulation of STAT protein import into nucleus(GO:2000366) |
| 0.4 | 1.7 | GO:0072308 | negative regulation by virus of viral protein levels in host cell(GO:0046725) negative regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072308) |
| 0.4 | 1.7 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.4 | 1.4 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.2 | 1.2 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.2 | 0.7 | GO:0002894 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.2 | 1.1 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) interferon-alpha biosynthetic process(GO:0045349) regulation of interferon-alpha biosynthetic process(GO:0045354) |
| 0.2 | 2.9 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.2 | 1.4 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.2 | 0.5 | GO:1903625 | negative regulation of DNA catabolic process(GO:1903625) regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 0.1 | 0.4 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.1 | 0.8 | GO:0019557 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.1 | 7.7 | GO:0032732 | positive regulation of interleukin-1 production(GO:0032732) |
| 0.1 | 0.4 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.1 | 3.2 | GO:0032461 | positive regulation of protein oligomerization(GO:0032461) |
| 0.1 | 0.3 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.1 | 0.3 | GO:0045080 | positive regulation of chemokine biosynthetic process(GO:0045080) |
| 0.1 | 0.9 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.9 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.1 | 1.2 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.1 | 1.7 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.2 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.0 | 0.2 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.6 | GO:0015866 | ADP transport(GO:0015866) |
| 0.0 | 0.1 | GO:1901340 | negative regulation of store-operated calcium channel activity(GO:1901340) |
| 0.0 | 0.6 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 0.0 | 0.4 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.5 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 2.4 | GO:0050918 | positive chemotaxis(GO:0050918) |
| 0.0 | 0.1 | GO:1903375 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) neural crest cell migration involved in sympathetic nervous system development(GO:1903045) facioacoustic ganglion development(GO:1903375) |
| 0.0 | 0.1 | GO:1905167 | positive regulation of lysosomal protein catabolic process(GO:1905167) |
| 0.0 | 0.1 | GO:0060481 | lobar bronchus epithelium development(GO:0060481) |
| 0.0 | 0.1 | GO:0051710 | regulation of cytolysis in other organism(GO:0051710) |
| 0.0 | 0.1 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.6 | GO:0060334 | regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.1 | GO:0044793 | negative regulation by host of viral process(GO:0044793) negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.0 | 0.2 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.3 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.0 | 1.2 | GO:0034394 | protein localization to cell surface(GO:0034394) |
| 0.0 | 0.1 | GO:0098912 | membrane depolarization during atrial cardiac muscle cell action potential(GO:0098912) |
| 0.0 | 0.3 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 | 0.5 | GO:0000717 | nucleotide-excision repair, DNA duplex unwinding(GO:0000717) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 2.9 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 0.4 | 2.8 | GO:0097179 | protease inhibitor complex(GO:0097179) |
| 0.1 | 2.8 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.1 | 0.5 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.2 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 1.0 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.6 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.3 | GO:0019774 | proteasome core complex, beta-subunit complex(GO:0019774) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0070996 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) type 1 melanocortin receptor binding(GO:0070996) |
| 0.3 | 0.8 | GO:0004397 | histidine ammonia-lyase activity(GO:0004397) |
| 0.2 | 2.8 | GO:0089720 | caspase binding(GO:0089720) |
| 0.2 | 1.2 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.1 | 0.4 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.1 | 0.9 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.6 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.1 | 5.1 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 0.5 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 0.5 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.1 | 0.4 | GO:0047787 | delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 0.1 | 3.0 | GO:0001848 | complement binding(GO:0001848) |
| 0.1 | 0.2 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.0 | 1.7 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.6 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.2 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.0 | 0.2 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.1 | GO:0016964 | alpha-2 macroglobulin receptor activity(GO:0016964) |
| 0.0 | 0.2 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 5.2 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 1.6 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 5.1 | GO:0005125 | cytokine activity(GO:0005125) |
| 0.0 | 3.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.0 | 0.4 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.3 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.7 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.1 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.2 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.1 | 2.9 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 4.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.1 | 2.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 1.8 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.7 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.5 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.1 | 3.0 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 1.9 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.1 | 1.4 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 2.6 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.6 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.4 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.5 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |