Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for CDX2
Z-value: 0.90
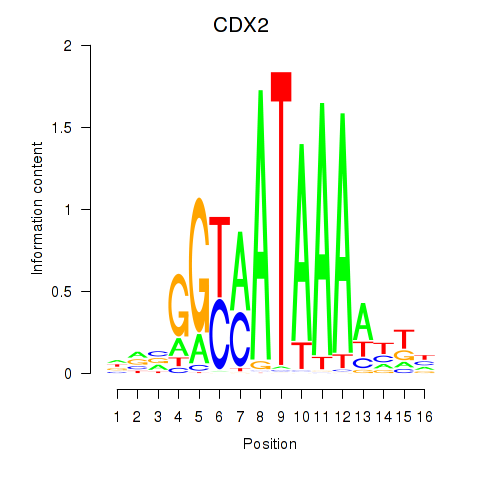
Transcription factors associated with CDX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CDX2
|
ENSG00000165556.9 | caudal type homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CDX2 | hg19_v2_chr13_-_28545276_28545276 | -0.74 | 2.6e-01 | Click! |
Activity profile of CDX2 motif
Sorted Z-values of CDX2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_39944025 | 0.72 |
ENST00000554328.1
ENST00000556620.1 ENST00000557197.1 |
RP11-111A21.1
|
RP11-111A21.1 |
| chr6_-_27782548 | 0.64 |
ENST00000333151.3
|
HIST1H2AJ
|
histone cluster 1, H2aj |
| chr17_-_46690839 | 0.47 |
ENST00000498634.2
|
HOXB8
|
homeobox B8 |
| chr17_+_58018269 | 0.44 |
ENST00000591035.1
|
RP11-178C3.1
|
Uncharacterized protein |
| chr4_-_140222358 | 0.44 |
ENST00000505036.1
ENST00000544855.1 ENST00000539002.1 |
NDUFC1
|
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr6_+_32812568 | 0.43 |
ENST00000414474.1
|
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr6_-_26189304 | 0.41 |
ENST00000340756.2
|
HIST1H4D
|
histone cluster 1, H4d |
| chr8_+_145215928 | 0.40 |
ENST00000528919.1
|
MROH1
|
maestro heat-like repeat family member 1 |
| chr6_+_26156551 | 0.37 |
ENST00000304218.3
|
HIST1H1E
|
histone cluster 1, H1e |
| chr14_+_56584414 | 0.35 |
ENST00000559044.1
|
PELI2
|
pellino E3 ubiquitin protein ligase family member 2 |
| chr9_-_130889990 | 0.33 |
ENST00000449878.1
|
PTGES2
|
prostaglandin E synthase 2 |
| chr2_-_32390801 | 0.32 |
ENST00000608489.1
|
RP11-563N4.1
|
RP11-563N4.1 |
| chr1_+_158979686 | 0.32 |
ENST00000368132.3
ENST00000295809.7 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr12_-_14924056 | 0.30 |
ENST00000539745.1
|
HIST4H4
|
histone cluster 4, H4 |
| chrM_+_10053 | 0.30 |
ENST00000361227.2
|
MT-ND3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr1_+_158979792 | 0.30 |
ENST00000359709.3
ENST00000430894.2 |
IFI16
|
interferon, gamma-inducible protein 16 |
| chr9_-_133814527 | 0.29 |
ENST00000451466.1
|
FIBCD1
|
fibrinogen C domain containing 1 |
| chr1_-_95846528 | 0.28 |
ENST00000427695.3
|
RP11-14O19.2
|
RP11-14O19.2 |
| chr4_+_186317133 | 0.28 |
ENST00000507753.1
|
ANKRD37
|
ankyrin repeat domain 37 |
| chr11_+_58390132 | 0.27 |
ENST00000361987.4
|
CNTF
|
ciliary neurotrophic factor |
| chr3_-_123710893 | 0.26 |
ENST00000467907.1
ENST00000459660.1 ENST00000495093.1 ENST00000460743.1 ENST00000405845.3 ENST00000484329.1 ENST00000479867.1 ENST00000496145.1 |
ROPN1
|
rhophilin associated tail protein 1 |
| chr19_-_48753028 | 0.25 |
ENST00000522431.1
|
CARD8
|
caspase recruitment domain family, member 8 |
| chr19_+_4402659 | 0.25 |
ENST00000301280.5
ENST00000585854.1 |
CHAF1A
|
chromatin assembly factor 1, subunit A (p150) |
| chr7_-_41742697 | 0.25 |
ENST00000242208.4
|
INHBA
|
inhibin, beta A |
| chr19_-_22018966 | 0.24 |
ENST00000599906.1
ENST00000354959.4 |
ZNF43
|
zinc finger protein 43 |
| chr9_+_100174232 | 0.24 |
ENST00000355295.4
|
TDRD7
|
tudor domain containing 7 |
| chr9_+_100174344 | 0.24 |
ENST00000422139.2
|
TDRD7
|
tudor domain containing 7 |
| chr19_-_10420459 | 0.24 |
ENST00000403352.1
ENST00000403903.3 |
ZGLP1
|
zinc finger, GATA-like protein 1 |
| chr17_+_61086917 | 0.24 |
ENST00000424789.2
ENST00000389520.4 |
TANC2
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr6_+_27775899 | 0.24 |
ENST00000358739.3
|
HIST1H2AI
|
histone cluster 1, H2ai |
| chr9_+_44868935 | 0.23 |
ENST00000448436.2
|
RP11-160N1.10
|
RP11-160N1.10 |
| chr5_+_61874696 | 0.23 |
ENST00000491184.2
|
LRRC70
|
leucine rich repeat containing 70 |
| chr16_+_14805546 | 0.22 |
ENST00000552140.1
|
NPIPA3
|
nuclear pore complex interacting protein family, member A3 |
| chr19_-_54618650 | 0.22 |
ENST00000391757.1
|
TFPT
|
TCF3 (E2A) fusion partner (in childhood Leukemia) |
| chr4_-_72649763 | 0.21 |
ENST00000513476.1
|
GC
|
group-specific component (vitamin D binding protein) |
| chr20_+_2276639 | 0.21 |
ENST00000381458.5
|
TGM3
|
transglutaminase 3 |
| chr9_-_133814455 | 0.21 |
ENST00000448616.1
|
FIBCD1
|
fibrinogen C domain containing 1 |
| chr8_-_73793975 | 0.20 |
ENST00000523881.1
|
RP11-1145L24.1
|
RP11-1145L24.1 |
| chr16_-_21431078 | 0.20 |
ENST00000458643.2
|
NPIPB3
|
nuclear pore complex interacting protein family, member B3 |
| chr3_+_107602030 | 0.20 |
ENST00000494231.1
|
LINC00636
|
long intergenic non-protein coding RNA 636 |
| chr6_-_133035185 | 0.19 |
ENST00000367928.4
|
VNN1
|
vanin 1 |
| chr12_+_54384370 | 0.19 |
ENST00000504315.1
|
HOXC6
|
homeobox C6 |
| chr9_+_137979506 | 0.19 |
ENST00000539529.1
ENST00000392991.4 ENST00000371793.3 |
OLFM1
|
olfactomedin 1 |
| chr5_+_121465207 | 0.19 |
ENST00000296600.4
|
ZNF474
|
zinc finger protein 474 |
| chr19_+_21264943 | 0.18 |
ENST00000597424.1
|
ZNF714
|
zinc finger protein 714 |
| chr22_+_17956618 | 0.18 |
ENST00000262608.8
|
CECR2
|
cat eye syndrome chromosome region, candidate 2 |
| chr2_-_122262593 | 0.18 |
ENST00000418989.1
|
CLASP1
|
cytoplasmic linker associated protein 1 |
| chr16_-_25122735 | 0.17 |
ENST00000563176.1
|
RP11-449H11.1
|
RP11-449H11.1 |
| chr19_-_54619006 | 0.17 |
ENST00000391759.1
|
TFPT
|
TCF3 (E2A) fusion partner (in childhood Leukemia) |
| chr7_+_6121296 | 0.16 |
ENST00000428901.1
|
AC004895.4
|
AC004895.4 |
| chr1_+_74701062 | 0.16 |
ENST00000326637.3
|
TNNI3K
|
TNNI3 interacting kinase |
| chr5_-_176433565 | 0.16 |
ENST00000428382.2
|
UIMC1
|
ubiquitin interaction motif containing 1 |
| chr19_-_6057282 | 0.15 |
ENST00000592281.1
|
RFX2
|
regulatory factor X, 2 (influences HLA class II expression) |
| chr16_-_15472151 | 0.15 |
ENST00000360151.4
ENST00000543801.1 |
NPIPA5
|
nuclear pore complex interacting protein family, member A5 |
| chr12_-_76377795 | 0.15 |
ENST00000552856.1
|
RP11-114H23.1
|
RP11-114H23.1 |
| chr7_+_35756092 | 0.14 |
ENST00000458087.3
|
AC018647.3
|
AC018647.3 |
| chr7_+_80275953 | 0.14 |
ENST00000538969.1
ENST00000544133.1 ENST00000433696.2 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr2_+_210444142 | 0.14 |
ENST00000360351.4
ENST00000361559.4 |
MAP2
|
microtubule-associated protein 2 |
| chr11_+_118938485 | 0.14 |
ENST00000300793.6
|
VPS11
|
vacuolar protein sorting 11 homolog (S. cerevisiae) |
| chr16_+_16472912 | 0.14 |
ENST00000530217.2
|
NPIPA7
|
nuclear pore complex interacting protein family, member A7 |
| chr3_+_125687987 | 0.14 |
ENST00000514116.1
ENST00000251776.4 ENST00000504401.1 ENST00000513830.1 ENST00000508088.1 |
ROPN1B
|
rhophilin associated tail protein 1B |
| chr8_+_38662960 | 0.14 |
ENST00000524193.1
|
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr11_-_4719072 | 0.13 |
ENST00000396950.3
ENST00000532598.1 |
OR51E2
|
olfactory receptor, family 51, subfamily E, member 2 |
| chrX_-_15619076 | 0.13 |
ENST00000252519.3
|
ACE2
|
angiotensin I converting enzyme 2 |
| chr7_-_130080977 | 0.13 |
ENST00000223208.5
|
CEP41
|
centrosomal protein 41kDa |
| chr5_+_61874562 | 0.13 |
ENST00000334994.5
ENST00000409534.1 |
LRRC70
IPO11
|
leucine rich repeat containing 70 importin 11 |
| chr17_-_39646116 | 0.13 |
ENST00000328119.6
|
KRT36
|
keratin 36 |
| chr4_-_156787425 | 0.13 |
ENST00000537611.2
|
ASIC5
|
acid-sensing (proton-gated) ion channel family member 5 |
| chr5_-_139937895 | 0.13 |
ENST00000336283.6
|
SRA1
|
steroid receptor RNA activator 1 |
| chr16_-_21663950 | 0.13 |
ENST00000268389.4
|
IGSF6
|
immunoglobulin superfamily, member 6 |
| chr6_+_134274354 | 0.12 |
ENST00000367869.1
|
TBPL1
|
TBP-like 1 |
| chr6_-_127840336 | 0.12 |
ENST00000525778.1
|
SOGA3
|
SOGA family member 3 |
| chr2_+_219081817 | 0.12 |
ENST00000315717.5
ENST00000420104.1 ENST00000295685.10 |
ARPC2
|
actin related protein 2/3 complex, subunit 2, 34kDa |
| chr2_+_210443993 | 0.12 |
ENST00000392193.1
|
MAP2
|
microtubule-associated protein 2 |
| chr7_+_35756186 | 0.12 |
ENST00000430518.1
|
AC018647.3
|
AC018647.3 |
| chr7_+_80275752 | 0.12 |
ENST00000419819.2
|
CD36
|
CD36 molecule (thrombospondin receptor) |
| chr19_+_54619125 | 0.12 |
ENST00000445811.1
ENST00000419967.1 ENST00000445124.1 ENST00000447810.1 |
PRPF31
|
pre-mRNA processing factor 31 |
| chr3_-_112127981 | 0.12 |
ENST00000486726.2
|
RP11-231E6.1
|
RP11-231E6.1 |
| chr4_+_129349188 | 0.12 |
ENST00000511497.1
|
RP11-420A23.1
|
RP11-420A23.1 |
| chr3_-_172241250 | 0.12 |
ENST00000420541.2
ENST00000241261.2 |
TNFSF10
|
tumor necrosis factor (ligand) superfamily, member 10 |
| chr19_+_13885252 | 0.12 |
ENST00000221576.4
|
C19orf53
|
chromosome 19 open reading frame 53 |
| chr1_-_95846556 | 0.12 |
ENST00000426881.2
|
RP11-14O19.2
|
RP11-14O19.2 |
| chr22_+_32455111 | 0.11 |
ENST00000543737.1
|
SLC5A1
|
solute carrier family 5 (sodium/glucose cotransporter), member 1 |
| chr2_-_217236750 | 0.11 |
ENST00000273067.4
|
MARCH4
|
membrane-associated ring finger (C3HC4) 4, E3 ubiquitin protein ligase |
| chr12_-_94673956 | 0.11 |
ENST00000551941.1
|
RP11-1105G2.3
|
Uncharacterized protein |
| chr16_+_71560023 | 0.11 |
ENST00000572450.1
|
CHST4
|
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 4 |
| chr11_-_65359947 | 0.11 |
ENST00000597463.1
|
AP001362.1
|
Uncharacterized protein |
| chr11_+_60048053 | 0.11 |
ENST00000337908.4
|
MS4A4A
|
membrane-spanning 4-domains, subfamily A, member 4A |
| chr2_-_169887827 | 0.11 |
ENST00000263817.6
|
ABCB11
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11 |
| chr14_-_38028689 | 0.11 |
ENST00000553425.1
|
RP11-356O9.2
|
RP11-356O9.2 |
| chr2_+_233415363 | 0.11 |
ENST00000409514.1
ENST00000409098.1 ENST00000409495.1 ENST00000409167.3 ENST00000409322.1 ENST00000409394.1 |
EIF4E2
|
eukaryotic translation initiation factor 4E family member 2 |
| chr1_+_46152886 | 0.11 |
ENST00000372025.4
|
TMEM69
|
transmembrane protein 69 |
| chr3_-_18480260 | 0.11 |
ENST00000454909.2
|
SATB1
|
SATB homeobox 1 |
| chr10_-_129691195 | 0.10 |
ENST00000368671.3
|
CLRN3
|
clarin 3 |
| chr5_-_138534071 | 0.10 |
ENST00000394817.2
|
SIL1
|
SIL1 nucleotide exchange factor |
| chr4_+_169633310 | 0.10 |
ENST00000510998.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr9_-_73029540 | 0.10 |
ENST00000377126.2
|
KLF9
|
Kruppel-like factor 9 |
| chr11_-_3663212 | 0.10 |
ENST00000397067.3
|
ART5
|
ADP-ribosyltransferase 5 |
| chr6_-_166582107 | 0.10 |
ENST00000296946.2
ENST00000461348.2 ENST00000366871.3 |
T
|
T, brachyury homolog (mouse) |
| chr14_+_52456193 | 0.09 |
ENST00000261700.3
|
C14orf166
|
chromosome 14 open reading frame 166 |
| chr5_+_136070614 | 0.09 |
ENST00000502421.1
|
CTB-1I21.1
|
CTB-1I21.1 |
| chr9_-_28670283 | 0.09 |
ENST00000379992.2
|
LINGO2
|
leucine rich repeat and Ig domain containing 2 |
| chr11_+_46366918 | 0.09 |
ENST00000528615.1
ENST00000395574.3 |
DGKZ
|
diacylglycerol kinase, zeta |
| chr1_+_154401791 | 0.09 |
ENST00000476006.1
|
IL6R
|
interleukin 6 receptor |
| chr20_+_4702548 | 0.09 |
ENST00000305817.2
|
PRND
|
prion protein 2 (dublet) |
| chr9_+_134065506 | 0.09 |
ENST00000483497.2
|
NUP214
|
nucleoporin 214kDa |
| chr1_-_54355430 | 0.09 |
ENST00000371399.1
ENST00000072644.1 ENST00000412288.1 |
YIPF1
|
Yip1 domain family, member 1 |
| chr17_-_72772425 | 0.09 |
ENST00000578822.1
|
NAT9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr7_+_63709496 | 0.09 |
ENST00000255746.4
|
ZNF679
|
zinc finger protein 679 |
| chrX_+_117957741 | 0.09 |
ENST00000310164.2
|
ZCCHC12
|
zinc finger, CCHC domain containing 12 |
| chr2_+_210444298 | 0.09 |
ENST00000445941.1
|
MAP2
|
microtubule-associated protein 2 |
| chr3_-_53925863 | 0.09 |
ENST00000541726.1
ENST00000495461.1 |
SELK
|
Selenoprotein K |
| chr18_+_55018044 | 0.09 |
ENST00000324000.3
|
ST8SIA3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
| chr17_-_72772462 | 0.08 |
ENST00000582870.1
ENST00000581136.1 ENST00000357814.3 ENST00000579218.1 ENST00000583476.1 ENST00000580301.1 ENST00000583757.1 ENST00000582524.1 |
NAT9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr14_+_35514323 | 0.08 |
ENST00000555211.1
|
FAM177A1
|
family with sequence similarity 177, member A1 |
| chr13_+_108921977 | 0.08 |
ENST00000430559.1
ENST00000375887.4 |
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b |
| chr10_+_47657581 | 0.08 |
ENST00000441923.2
|
ANTXRL
|
anthrax toxin receptor-like |
| chr19_-_48759119 | 0.08 |
ENST00000522889.1
ENST00000520753.1 ENST00000519940.1 ENST00000519332.1 ENST00000521437.1 ENST00000520007.1 ENST00000521613.1 |
CARD8
|
caspase recruitment domain family, member 8 |
| chr12_-_62586543 | 0.08 |
ENST00000416284.3
|
FAM19A2
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A2 |
| chr1_+_24645807 | 0.08 |
ENST00000361548.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr1_+_73771844 | 0.08 |
ENST00000440762.1
ENST00000444827.1 ENST00000415686.1 ENST00000411903.1 |
RP4-598G3.1
|
RP4-598G3.1 |
| chr2_-_113810444 | 0.08 |
ENST00000259213.4
ENST00000327407.2 |
IL36B
|
interleukin 36, beta |
| chr15_-_98417780 | 0.08 |
ENST00000503874.3
|
LINC00923
|
long intergenic non-protein coding RNA 923 |
| chr3_+_63953415 | 0.08 |
ENST00000484332.1
|
ATXN7
|
ataxin 7 |
| chr6_+_125304502 | 0.08 |
ENST00000519799.1
ENST00000368414.2 ENST00000359704.2 |
RNF217
|
ring finger protein 217 |
| chr7_+_80275621 | 0.08 |
ENST00000426978.1
ENST00000432207.1 |
CD36
|
CD36 molecule (thrombospondin receptor) |
| chrX_+_65384052 | 0.08 |
ENST00000336279.5
ENST00000458621.1 |
HEPH
|
hephaestin |
| chr8_-_13372395 | 0.08 |
ENST00000276297.4
ENST00000511869.1 |
DLC1
|
deleted in liver cancer 1 |
| chr7_+_141811539 | 0.08 |
ENST00000550469.2
ENST00000477922.3 |
RP11-1220K2.2
|
Putative inactive maltase-glucoamylase-like protein LOC93432 |
| chr12_+_14561422 | 0.08 |
ENST00000541056.1
|
ATF7IP
|
activating transcription factor 7 interacting protein |
| chr5_+_176731572 | 0.08 |
ENST00000503853.1
|
PRELID1
|
PRELI domain containing 1 |
| chr17_-_76921459 | 0.07 |
ENST00000262768.7
|
TIMP2
|
TIMP metallopeptidase inhibitor 2 |
| chr2_-_224467002 | 0.07 |
ENST00000421386.1
ENST00000433889.1 |
SCG2
|
secretogranin II |
| chr17_-_80797886 | 0.07 |
ENST00000572562.1
|
ZNF750
|
zinc finger protein 750 |
| chr13_-_103719196 | 0.07 |
ENST00000245312.3
|
SLC10A2
|
solute carrier family 10 (sodium/bile acid cotransporter), member 2 |
| chr16_-_20339123 | 0.07 |
ENST00000381360.5
|
GP2
|
glycoprotein 2 (zymogen granule membrane) |
| chr6_+_25754927 | 0.07 |
ENST00000377905.4
ENST00000439485.2 |
SLC17A4
|
solute carrier family 17, member 4 |
| chr22_-_30642782 | 0.07 |
ENST00000249075.3
|
LIF
|
leukemia inhibitory factor |
| chrX_+_65384182 | 0.07 |
ENST00000441993.2
ENST00000419594.1 |
HEPH
|
hephaestin |
| chr1_+_151739131 | 0.07 |
ENST00000400999.1
|
OAZ3
|
ornithine decarboxylase antizyme 3 |
| chr14_+_52456327 | 0.07 |
ENST00000556760.1
|
C14orf166
|
chromosome 14 open reading frame 166 |
| chr20_+_56964169 | 0.07 |
ENST00000475243.1
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr4_+_146539415 | 0.07 |
ENST00000281317.5
|
MMAA
|
methylmalonic aciduria (cobalamin deficiency) cblA type |
| chr1_+_40942887 | 0.06 |
ENST00000372706.1
|
ZFP69
|
ZFP69 zinc finger protein |
| chr7_+_57509877 | 0.06 |
ENST00000420713.1
|
ZNF716
|
zinc finger protein 716 |
| chr5_-_177180297 | 0.06 |
ENST00000504518.1
|
FAM153A
|
family with sequence similarity 153, member A |
| chr19_-_23869999 | 0.06 |
ENST00000601935.1
ENST00000359788.4 ENST00000600313.1 ENST00000596211.1 ENST00000599168.1 |
ZNF675
|
zinc finger protein 675 |
| chr18_+_61445205 | 0.06 |
ENST00000431370.1
|
SERPINB7
|
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr11_-_60674037 | 0.06 |
ENST00000541371.1
ENST00000227524.4 |
PRPF19
|
pre-mRNA processing factor 19 |
| chr6_+_53948328 | 0.06 |
ENST00000370876.2
|
MLIP
|
muscular LMNA-interacting protein |
| chr1_+_52682052 | 0.06 |
ENST00000371591.1
|
ZFYVE9
|
zinc finger, FYVE domain containing 9 |
| chr16_-_20338748 | 0.06 |
ENST00000575582.1
ENST00000341642.5 ENST00000381362.4 ENST00000572347.1 ENST00000572478.1 ENST00000302555.5 |
GP2
|
glycoprotein 2 (zymogen granule membrane) |
| chr13_+_108922228 | 0.06 |
ENST00000542136.1
|
TNFSF13B
|
tumor necrosis factor (ligand) superfamily, member 13b |
| chr1_-_204135450 | 0.05 |
ENST00000272190.8
ENST00000367195.2 |
REN
|
renin |
| chr15_+_22736484 | 0.05 |
ENST00000560659.2
|
GOLGA6L1
|
golgin A6 family-like 1 |
| chr5_-_159766528 | 0.05 |
ENST00000505287.2
|
CCNJL
|
cyclin J-like |
| chr2_-_95787738 | 0.05 |
ENST00000272418.2
|
MRPS5
|
mitochondrial ribosomal protein S5 |
| chr16_+_71560154 | 0.05 |
ENST00000539698.3
|
CHST4
|
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 4 |
| chr1_+_24645865 | 0.05 |
ENST00000342072.4
|
GRHL3
|
grainyhead-like 3 (Drosophila) |
| chr7_-_26578407 | 0.05 |
ENST00000242109.3
|
KIAA0087
|
KIAA0087 |
| chr3_+_15045419 | 0.05 |
ENST00000406272.2
|
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr12_-_95774672 | 0.05 |
ENST00000549961.1
|
RP11-167N24.6
|
Uncharacterized protein |
| chr2_-_176046391 | 0.05 |
ENST00000392541.3
ENST00000409194.1 |
ATP5G3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9) |
| chrX_+_140982452 | 0.05 |
ENST00000544766.1
|
MAGEC3
|
melanoma antigen family C, 3 |
| chr1_-_23495340 | 0.05 |
ENST00000418342.1
|
LUZP1
|
leucine zipper protein 1 |
| chr11_+_22688150 | 0.05 |
ENST00000454584.2
|
GAS2
|
growth arrest-specific 2 |
| chr20_+_30102231 | 0.05 |
ENST00000335574.5
ENST00000340852.5 ENST00000398174.3 ENST00000376127.3 ENST00000344042.5 |
HM13
|
histocompatibility (minor) 13 |
| chr12_+_56546223 | 0.05 |
ENST00000550443.1
ENST00000207437.5 |
MYL6B
|
myosin, light chain 6B, alkali, smooth muscle and non-muscle |
| chr10_-_96829246 | 0.05 |
ENST00000371270.3
ENST00000535898.1 ENST00000539050.1 |
CYP2C8
|
cytochrome P450, family 2, subfamily C, polypeptide 8 |
| chrX_-_135962923 | 0.05 |
ENST00000565438.1
|
RBMX
|
RNA binding motif protein, X-linked |
| chr5_-_148442584 | 0.05 |
ENST00000394358.2
ENST00000512049.1 |
SH3TC2
|
SH3 domain and tetratricopeptide repeats 2 |
| chr12_-_76879852 | 0.05 |
ENST00000548341.1
|
OSBPL8
|
oxysterol binding protein-like 8 |
| chr14_-_60097524 | 0.05 |
ENST00000342503.4
|
RTN1
|
reticulon 1 |
| chr6_+_158733692 | 0.05 |
ENST00000367094.2
ENST00000367097.3 |
TULP4
|
tubby like protein 4 |
| chr1_-_43919346 | 0.04 |
ENST00000372430.3
ENST00000372433.1 ENST00000372434.1 ENST00000486909.1 |
HYI
|
hydroxypyruvate isomerase (putative) |
| chr2_-_216878305 | 0.04 |
ENST00000263268.6
|
MREG
|
melanoregulin |
| chr1_-_106161540 | 0.04 |
ENST00000420901.1
ENST00000610126.1 ENST00000435253.2 |
RP11-251P6.1
|
RP11-251P6.1 |
| chr15_+_22736246 | 0.04 |
ENST00000316397.3
|
GOLGA6L1
|
golgin A6 family-like 1 |
| chrX_-_131353461 | 0.04 |
ENST00000370874.1
|
RAP2C
|
RAP2C, member of RAS oncogene family |
| chr15_+_78830023 | 0.04 |
ENST00000599596.1
|
AC027228.1
|
HCG2005369; Uncharacterized protein |
| chr3_-_38992052 | 0.04 |
ENST00000302328.3
ENST00000450244.1 ENST00000444237.2 |
SCN11A
|
sodium channel, voltage-gated, type XI, alpha subunit |
| chr1_-_85100703 | 0.04 |
ENST00000370624.1
|
C1orf180
|
chromosome 1 open reading frame 180 |
| chr12_+_65996599 | 0.04 |
ENST00000539116.1
ENST00000541391.1 |
RP11-221N13.3
|
RP11-221N13.3 |
| chr3_+_185046676 | 0.04 |
ENST00000428617.1
ENST00000443863.1 |
MAP3K13
|
mitogen-activated protein kinase kinase kinase 13 |
| chr7_+_138915102 | 0.04 |
ENST00000486663.1
|
UBN2
|
ubinuclein 2 |
| chr2_+_27440229 | 0.04 |
ENST00000264705.4
ENST00000403525.1 |
CAD
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr12_-_118796971 | 0.04 |
ENST00000542902.1
|
TAOK3
|
TAO kinase 3 |
| chr16_+_25123041 | 0.04 |
ENST00000399069.3
ENST00000380966.4 |
LCMT1
|
leucine carboxyl methyltransferase 1 |
| chr10_-_127505167 | 0.04 |
ENST00000368786.1
|
UROS
|
uroporphyrinogen III synthase |
| chr12_+_1800179 | 0.04 |
ENST00000357103.4
|
ADIPOR2
|
adiponectin receptor 2 |
| chr22_+_41956767 | 0.04 |
ENST00000306149.7
|
CSDC2
|
cold shock domain containing C2, RNA binding |
| chr3_+_143690640 | 0.04 |
ENST00000315691.3
|
C3orf58
|
chromosome 3 open reading frame 58 |
| chr16_-_25122785 | 0.04 |
ENST00000563962.1
ENST00000569920.1 |
RP11-449H11.1
|
RP11-449H11.1 |
| chr8_+_128426535 | 0.04 |
ENST00000465342.2
|
POU5F1B
|
POU class 5 homeobox 1B |
| chr19_-_48752812 | 0.04 |
ENST00000359009.4
|
CARD8
|
caspase recruitment domain family, member 8 |
| chrX_+_65382381 | 0.04 |
ENST00000519389.1
|
HEPH
|
hephaestin |
| chr16_+_68119764 | 0.04 |
ENST00000570212.1
ENST00000562926.1 |
NFATC3
|
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3 |
| chr4_-_186317034 | 0.04 |
ENST00000505916.1
|
LRP2BP
|
LRP2 binding protein |
| chr3_-_9921934 | 0.04 |
ENST00000423850.1
|
CIDEC
|
cell death-inducing DFFA-like effector c |
| chr13_+_24144509 | 0.03 |
ENST00000248484.4
|
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
Network of associatons between targets according to the STRING database.
First level regulatory network of CDX2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0060279 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.1 | 0.3 | GO:0046668 | regulation of retinal cell programmed cell death(GO:0046668) |
| 0.1 | 0.4 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.3 | GO:2000332 | blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 0.4 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.0 | 0.1 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.0 | 0.2 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.1 | GO:1900133 | regulation of renin secretion into blood stream(GO:1900133) |
| 0.0 | 0.1 | GO:0002035 | brain renin-angiotensin system(GO:0002035) angiotensin-mediated drinking behavior(GO:0003051) |
| 0.0 | 0.4 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.0 | 0.3 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 0.1 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.0 | 0.1 | GO:0000349 | generation of catalytic spliceosome for first transesterification step(GO:0000349) |
| 0.0 | 0.1 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.0 | 0.2 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.5 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.2 | GO:0003190 | atrioventricular valve formation(GO:0003190) |
| 0.0 | 0.1 | GO:0021503 | neural fold bending(GO:0021503) |
| 0.0 | 0.0 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.0 | 0.1 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.3 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.1 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.0 | 0.1 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.2 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.1 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.0 | 0.0 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.0 | 0.1 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 0.0 | 0.1 | GO:2000643 | positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.0 | 0.2 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.1 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.0 | 0.5 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.0 | 0.2 | GO:2000400 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 0.1 | GO:1901857 | positive regulation of cellular respiration(GO:1901857) |
| 0.0 | 0.6 | GO:0032731 | positive regulation of interleukin-1 beta production(GO:0032731) |
| 0.0 | 0.1 | GO:0018095 | protein polyglutamylation(GO:0018095) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.1 | 0.3 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.1 | 0.4 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.3 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.1 | 0.4 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.2 | GO:0031592 | centrosomal corona(GO:0031592) |
| 0.0 | 0.1 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.4 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.5 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.4 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.0 | 0.1 | GO:0071006 | U2-type catalytic step 1 spliceosome(GO:0071006) |
| 0.0 | 0.6 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.4 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.0 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.2 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.2 | GO:0031010 | ISWI-type complex(GO:0031010) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1902271 | D3 vitamins binding(GO:1902271) |
| 0.1 | 0.3 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.1 | 0.4 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.1 | 0.4 | GO:0032564 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.1 | 0.3 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.2 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.1 | GO:0015432 | bile acid-exporting ATPase activity(GO:0015432) |
| 0.0 | 0.4 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.2 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.5 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.3 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.4 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.2 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.2 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.1 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.4 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.2 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.1 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.4 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.0 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.0 | 0.0 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.0 | 0.1 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.0 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.4 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.4 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |