Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
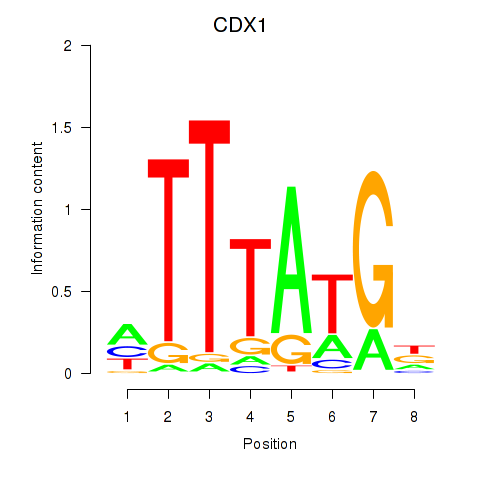
Results for CDX1
Z-value: 1.18
Transcription factors associated with CDX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CDX1
|
ENSG00000113722.12 | caudal type homeobox 1 |
Activity profile of CDX1 motif
Sorted Z-values of CDX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_90712520 | 1.45 |
ENST00000224784.6
|
ACTA2
|
actin, alpha 2, smooth muscle, aorta |
| chr14_+_88490894 | 0.96 |
ENST00000556033.1
ENST00000553929.1 ENST00000555996.1 ENST00000556673.1 ENST00000557339.1 ENST00000556684.1 |
RP11-300J18.3
|
long intergenic non-protein coding RNA 1146 |
| chr2_-_44550441 | 0.94 |
ENST00000420756.1
ENST00000444696.1 |
PREPL
|
prolyl endopeptidase-like |
| chr3_-_148939598 | 0.91 |
ENST00000455472.3
|
CP
|
ceruloplasmin (ferroxidase) |
| chr1_+_241695670 | 0.86 |
ENST00000366557.4
|
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr17_-_57229155 | 0.86 |
ENST00000584089.1
|
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr4_-_110723335 | 0.80 |
ENST00000394634.2
|
CFI
|
complement factor I |
| chr1_-_197036364 | 0.76 |
ENST00000367412.1
|
F13B
|
coagulation factor XIII, B polypeptide |
| chr2_+_161993465 | 0.75 |
ENST00000457476.1
|
TANK
|
TRAF family member-associated NFKB activator |
| chr7_-_16840820 | 0.75 |
ENST00000450569.1
|
AGR2
|
anterior gradient 2 |
| chr7_-_35013217 | 0.74 |
ENST00000446375.1
|
DPY19L1
|
dpy-19-like 1 (C. elegans) |
| chr3_+_149192475 | 0.68 |
ENST00000465758.1
|
TM4SF4
|
transmembrane 4 L six family member 4 |
| chr4_-_110723194 | 0.68 |
ENST00000394635.3
|
CFI
|
complement factor I |
| chr21_+_17909594 | 0.64 |
ENST00000441820.1
ENST00000602280.1 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr1_+_212475148 | 0.63 |
ENST00000537030.3
|
PPP2R5A
|
protein phosphatase 2, regulatory subunit B', alpha |
| chr16_+_2014993 | 0.63 |
ENST00000564014.1
|
SNHG9
|
small nucleolar RNA host gene 9 (non-protein coding) |
| chr10_-_69597915 | 0.62 |
ENST00000225171.2
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr18_+_3466248 | 0.62 |
ENST00000581029.1
ENST00000581442.1 ENST00000579007.1 |
RP11-838N2.4
|
RP11-838N2.4 |
| chr15_-_45670924 | 0.60 |
ENST00000396659.3
|
GATM
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr9_-_69229650 | 0.58 |
ENST00000416428.1
|
CBWD6
|
COBW domain containing 6 |
| chr19_+_38880695 | 0.57 |
ENST00000587947.1
ENST00000338502.4 |
SPRED3
|
sprouty-related, EVH1 domain containing 3 |
| chr6_+_26087509 | 0.57 |
ENST00000397022.3
ENST00000353147.5 ENST00000352392.4 ENST00000349999.4 ENST00000317896.7 ENST00000357618.5 ENST00000470149.1 ENST00000336625.8 ENST00000461397.1 ENST00000488199.1 |
HFE
|
hemochromatosis |
| chr3_-_149095652 | 0.57 |
ENST00000305366.3
|
TM4SF1
|
transmembrane 4 L six family member 1 |
| chrX_-_80377118 | 0.53 |
ENST00000373250.3
|
HMGN5
|
high mobility group nucleosome binding domain 5 |
| chr10_-_69597828 | 0.52 |
ENST00000339758.7
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr1_-_111970353 | 0.51 |
ENST00000369732.3
|
OVGP1
|
oviductal glycoprotein 1, 120kDa |
| chr10_-_69597810 | 0.51 |
ENST00000483798.2
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr12_-_91573132 | 0.50 |
ENST00000550563.1
ENST00000546370.1 |
DCN
|
decorin |
| chr9_-_3469181 | 0.50 |
ENST00000366116.2
|
AL365202.1
|
Uncharacterized protein |
| chr8_-_95220775 | 0.49 |
ENST00000441892.2
ENST00000521491.1 ENST00000027335.3 |
CDH17
|
cadherin 17, LI cadherin (liver-intestine) |
| chr5_+_75904950 | 0.49 |
ENST00000502745.1
|
IQGAP2
|
IQ motif containing GTPase activating protein 2 |
| chr9_+_125795788 | 0.49 |
ENST00000373643.5
|
RABGAP1
|
RAB GTPase activating protein 1 |
| chr17_-_57232596 | 0.49 |
ENST00000581068.1
ENST00000330137.7 |
SKA2
|
spindle and kinetochore associated complex subunit 2 |
| chr7_-_7679633 | 0.49 |
ENST00000401447.1
|
RPA3
|
replication protein A3, 14kDa |
| chr6_+_63921351 | 0.48 |
ENST00000370659.1
|
FKBP1C
|
FK506 binding protein 1C |
| chr4_-_110723134 | 0.47 |
ENST00000510800.1
ENST00000512148.1 |
CFI
|
complement factor I |
| chr2_+_33359687 | 0.46 |
ENST00000402934.1
ENST00000404525.1 ENST00000407925.1 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr6_+_126661253 | 0.45 |
ENST00000368326.1
ENST00000368325.1 ENST00000368328.4 |
CENPW
|
centromere protein W |
| chr5_+_95997918 | 0.44 |
ENST00000395812.2
ENST00000395813.1 ENST00000359176.4 ENST00000325674.7 |
CAST
|
calpastatin |
| chr2_+_33359646 | 0.43 |
ENST00000390003.4
ENST00000418533.2 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr12_-_371994 | 0.41 |
ENST00000343164.4
ENST00000436453.1 ENST00000445055.2 ENST00000546319.1 |
SLC6A13
|
solute carrier family 6 (neurotransmitter transporter), member 13 |
| chr21_-_35773370 | 0.41 |
ENST00000410005.1
|
AP000322.54
|
chromosome 21 open reading frame 140 |
| chr10_-_126480381 | 0.40 |
ENST00000368836.2
|
METTL10
|
methyltransferase like 10 |
| chr2_+_132479948 | 0.40 |
ENST00000355171.4
|
C2orf27A
|
chromosome 2 open reading frame 27A |
| chr2_+_242289502 | 0.38 |
ENST00000451310.1
|
SEPT2
|
septin 2 |
| chr20_-_17539456 | 0.38 |
ENST00000544874.1
ENST00000377868.2 |
BFSP1
|
beaded filament structural protein 1, filensin |
| chr9_-_107754034 | 0.37 |
ENST00000457720.1
|
RP11-217B7.3
|
RP11-217B7.3 |
| chr11_+_107992243 | 0.37 |
ENST00000265838.4
ENST00000299355.6 |
ACAT1
|
acetyl-CoA acetyltransferase 1 |
| chr1_-_207095324 | 0.36 |
ENST00000530505.1
ENST00000367091.3 ENST00000442471.2 |
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr14_+_58797974 | 0.36 |
ENST00000417477.2
|
ARID4A
|
AT rich interactive domain 4A (RBP1-like) |
| chr8_-_103876383 | 0.36 |
ENST00000347770.4
|
AZIN1
|
antizyme inhibitor 1 |
| chr6_-_43423308 | 0.36 |
ENST00000372485.1
ENST00000372488.3 |
DLK2
|
delta-like 2 homolog (Drosophila) |
| chrY_+_14958970 | 0.36 |
ENST00000453031.1
|
USP9Y
|
ubiquitin specific peptidase 9, Y-linked |
| chr15_-_55700457 | 0.35 |
ENST00000442196.3
ENST00000563171.1 ENST00000425574.3 |
CCPG1
|
cell cycle progression 1 |
| chr9_-_97356075 | 0.35 |
ENST00000375337.3
|
FBP2
|
fructose-1,6-bisphosphatase 2 |
| chr17_-_48943706 | 0.35 |
ENST00000499247.2
|
TOB1
|
transducer of ERBB2, 1 |
| chr15_+_89164520 | 0.35 |
ENST00000332810.3
|
AEN
|
apoptosis enhancing nuclease |
| chr15_+_69857515 | 0.35 |
ENST00000559477.1
|
RP11-279F6.1
|
RP11-279F6.1 |
| chr17_+_57233087 | 0.34 |
ENST00000578777.1
ENST00000577457.1 ENST00000582995.1 |
PRR11
|
proline rich 11 |
| chr10_-_38146965 | 0.34 |
ENST00000395873.3
ENST00000357328.4 ENST00000395874.2 |
ZNF248
|
zinc finger protein 248 |
| chr1_-_15911510 | 0.34 |
ENST00000375826.3
|
AGMAT
|
agmatine ureohydrolase (agmatinase) |
| chr18_-_29264467 | 0.34 |
ENST00000383131.3
ENST00000237019.7 |
B4GALT6
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 |
| chr14_+_102276192 | 0.33 |
ENST00000557714.1
|
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr1_+_150254936 | 0.33 |
ENST00000447007.1
ENST00000369095.1 ENST00000369094.1 |
C1orf51
|
chromosome 1 open reading frame 51 |
| chr2_+_196521458 | 0.32 |
ENST00000409086.3
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr15_-_45670717 | 0.32 |
ENST00000558163.1
ENST00000558336.1 |
GATM
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr6_+_26087646 | 0.32 |
ENST00000309234.6
|
HFE
|
hemochromatosis |
| chr22_+_25595817 | 0.32 |
ENST00000215855.2
ENST00000404334.1 |
CRYBB3
|
crystallin, beta B3 |
| chr21_+_17214724 | 0.31 |
ENST00000449491.1
|
USP25
|
ubiquitin specific peptidase 25 |
| chr18_+_61575200 | 0.31 |
ENST00000238508.3
|
SERPINB10
|
serpin peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr16_+_56672571 | 0.31 |
ENST00000290705.8
|
MT1A
|
metallothionein 1A |
| chr12_-_76461249 | 0.31 |
ENST00000551524.1
|
NAP1L1
|
nucleosome assembly protein 1-like 1 |
| chr12_-_90049878 | 0.31 |
ENST00000359142.3
|
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr2_+_161993412 | 0.31 |
ENST00000259075.2
ENST00000432002.1 |
TANK
|
TRAF family member-associated NFKB activator |
| chr17_-_56492989 | 0.30 |
ENST00000583753.1
|
RNF43
|
ring finger protein 43 |
| chr7_+_23210760 | 0.30 |
ENST00000366347.4
|
AC005082.1
|
Uncharacterized protein |
| chr8_-_125551278 | 0.30 |
ENST00000519232.1
ENST00000523888.1 ENST00000522810.1 ENST00000519548.1 ENST00000517678.1 ENST00000605953.1 ENST00000276692.6 |
TATDN1
|
TatD DNase domain containing 1 |
| chr8_+_76452097 | 0.30 |
ENST00000396423.2
|
HNF4G
|
hepatocyte nuclear factor 4, gamma |
| chr15_-_83474806 | 0.30 |
ENST00000541889.1
ENST00000334574.8 ENST00000561368.1 |
FSD2
|
fibronectin type III and SPRY domain containing 2 |
| chr1_-_40042416 | 0.30 |
ENST00000372857.3
ENST00000372856.3 ENST00000531243.2 ENST00000451091.2 |
PABPC4
|
poly(A) binding protein, cytoplasmic 4 (inducible form) |
| chr12_+_16500037 | 0.29 |
ENST00000536371.1
ENST00000010404.2 |
MGST1
|
microsomal glutathione S-transferase 1 |
| chr13_+_53030107 | 0.29 |
ENST00000490903.1
ENST00000480747.1 |
CKAP2
|
cytoskeleton associated protein 2 |
| chr11_-_4719072 | 0.29 |
ENST00000396950.3
ENST00000532598.1 |
OR51E2
|
olfactory receptor, family 51, subfamily E, member 2 |
| chr9_+_124088860 | 0.29 |
ENST00000373806.1
|
GSN
|
gelsolin |
| chr8_+_35649365 | 0.29 |
ENST00000437887.1
|
AC012215.1
|
Uncharacterized protein |
| chr19_+_46367518 | 0.28 |
ENST00000302177.2
|
FOXA3
|
forkhead box A3 |
| chr7_+_90012986 | 0.28 |
ENST00000416322.1
|
CLDN12
|
claudin 12 |
| chr17_+_57232690 | 0.28 |
ENST00000262293.4
|
PRR11
|
proline rich 11 |
| chr3_-_96337000 | 0.28 |
ENST00000600213.2
|
MTRNR2L12
|
MT-RNR2-like 12 (pseudogene) |
| chr16_-_18887627 | 0.28 |
ENST00000563235.1
|
SMG1
|
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr7_+_151722712 | 0.28 |
ENST00000430044.2
ENST00000431668.1 ENST00000446096.1 ENST00000452146.2 ENST00000423337.1 |
GALNT11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
| chr12_+_28410128 | 0.28 |
ENST00000381259.1
ENST00000381256.1 |
CCDC91
|
coiled-coil domain containing 91 |
| chr1_-_193075180 | 0.28 |
ENST00000367440.3
|
GLRX2
|
glutaredoxin 2 |
| chr5_+_75904918 | 0.27 |
ENST00000514001.1
ENST00000396234.3 ENST00000509074.1 |
IQGAP2
|
IQ motif containing GTPase activating protein 2 |
| chr8_-_117886955 | 0.27 |
ENST00000297338.2
|
RAD21
|
RAD21 homolog (S. pombe) |
| chr13_-_114107839 | 0.27 |
ENST00000375418.3
|
ADPRHL1
|
ADP-ribosylhydrolase like 1 |
| chr18_+_47901408 | 0.27 |
ENST00000398452.2
ENST00000417656.2 ENST00000488454.1 ENST00000494518.1 |
SKA1
|
spindle and kinetochore associated complex subunit 1 |
| chr3_-_193096600 | 0.27 |
ENST00000446087.1
ENST00000342358.4 |
ATP13A5
|
ATPase type 13A5 |
| chr16_-_11876408 | 0.27 |
ENST00000396516.2
|
ZC3H7A
|
zinc finger CCCH-type containing 7A |
| chr8_+_93895865 | 0.26 |
ENST00000391681.1
|
AC117834.1
|
AC117834.1 |
| chr11_+_73003824 | 0.26 |
ENST00000538328.1
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr3_+_45636219 | 0.26 |
ENST00000273317.4
|
LIMD1
|
LIM domains containing 1 |
| chr2_-_224467093 | 0.26 |
ENST00000305409.2
|
SCG2
|
secretogranin II |
| chr10_+_112327425 | 0.26 |
ENST00000361804.4
|
SMC3
|
structural maintenance of chromosomes 3 |
| chr17_+_41005283 | 0.26 |
ENST00000592999.1
|
AOC3
|
amine oxidase, copper containing 3 |
| chrX_-_45629661 | 0.25 |
ENST00000602507.1
ENST00000602461.1 |
RP6-99M1.2
|
RP6-99M1.2 |
| chr5_+_96079240 | 0.25 |
ENST00000515663.1
|
CAST
|
calpastatin |
| chr7_+_37723450 | 0.25 |
ENST00000447769.1
|
GPR141
|
G protein-coupled receptor 141 |
| chr7_-_11871815 | 0.25 |
ENST00000423059.4
|
THSD7A
|
thrombospondin, type I, domain containing 7A |
| chr18_+_47901365 | 0.25 |
ENST00000285116.3
|
SKA1
|
spindle and kinetochore associated complex subunit 1 |
| chr5_+_115177178 | 0.24 |
ENST00000316788.7
|
AP3S1
|
adaptor-related protein complex 3, sigma 1 subunit |
| chr11_-_62439727 | 0.24 |
ENST00000528862.1
|
C11orf48
|
chromosome 11 open reading frame 48 |
| chr14_+_24407940 | 0.24 |
ENST00000354854.1
|
DHRS4-AS1
|
DHRS4-AS1 |
| chr5_+_138210919 | 0.24 |
ENST00000522013.1
ENST00000520260.1 ENST00000523298.1 ENST00000520865.1 ENST00000519634.1 ENST00000517533.1 ENST00000523685.1 ENST00000519768.1 ENST00000517656.1 ENST00000521683.1 ENST00000521640.1 ENST00000519116.1 |
CTNNA1
|
catenin (cadherin-associated protein), alpha 1, 102kDa |
| chr18_-_68004529 | 0.23 |
ENST00000578633.1
|
RP11-484N16.1
|
RP11-484N16.1 |
| chr6_+_76599809 | 0.23 |
ENST00000430435.1
|
MYO6
|
myosin VI |
| chr13_+_73302047 | 0.23 |
ENST00000377814.2
ENST00000377815.3 ENST00000390667.5 |
BORA
|
bora, aurora kinase A activator |
| chr6_+_30689350 | 0.23 |
ENST00000330914.3
|
TUBB
|
tubulin, beta class I |
| chr1_+_84630053 | 0.23 |
ENST00000394838.2
ENST00000370682.3 ENST00000432111.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr15_+_52121822 | 0.22 |
ENST00000558455.1
ENST00000308580.7 |
TMOD3
|
tropomodulin 3 (ubiquitous) |
| chr10_-_52645416 | 0.22 |
ENST00000374001.2
ENST00000373997.3 ENST00000373995.3 ENST00000282641.2 ENST00000395495.1 ENST00000414883.1 |
A1CF
|
APOBEC1 complementation factor |
| chr1_-_68299130 | 0.22 |
ENST00000370982.3
|
GNG12
|
guanine nucleotide binding protein (G protein), gamma 12 |
| chr12_-_31744031 | 0.22 |
ENST00000389082.5
|
DENND5B
|
DENN/MADD domain containing 5B |
| chr5_-_76916396 | 0.22 |
ENST00000509971.1
|
WDR41
|
WD repeat domain 41 |
| chr9_-_21335240 | 0.22 |
ENST00000537938.1
|
KLHL9
|
kelch-like family member 9 |
| chr8_+_19536083 | 0.22 |
ENST00000519803.1
|
RP11-1105O14.1
|
RP11-1105O14.1 |
| chr15_-_55700522 | 0.22 |
ENST00000564092.1
ENST00000310958.6 |
CCPG1
|
cell cycle progression 1 |
| chr11_+_28129795 | 0.22 |
ENST00000406787.3
ENST00000342303.5 ENST00000403099.1 ENST00000407364.3 |
METTL15
|
methyltransferase like 15 |
| chr18_+_20513782 | 0.22 |
ENST00000399722.2
ENST00000399725.2 ENST00000399721.2 ENST00000583594.1 |
RBBP8
|
retinoblastoma binding protein 8 |
| chr9_-_21335356 | 0.21 |
ENST00000359039.4
|
KLHL9
|
kelch-like family member 9 |
| chr3_-_197025447 | 0.21 |
ENST00000346964.2
ENST00000357674.4 ENST00000314062.3 ENST00000448528.2 ENST00000419553.1 |
DLG1
|
discs, large homolog 1 (Drosophila) |
| chr1_+_17559776 | 0.21 |
ENST00000537499.1
ENST00000413717.2 ENST00000536552.1 |
PADI1
|
peptidyl arginine deiminase, type I |
| chr13_+_78109884 | 0.21 |
ENST00000377246.3
ENST00000349847.3 |
SCEL
|
sciellin |
| chr8_+_107738240 | 0.21 |
ENST00000449762.2
ENST00000297447.6 |
OXR1
|
oxidation resistance 1 |
| chr19_+_18530184 | 0.21 |
ENST00000601357.2
|
SSBP4
|
single stranded DNA binding protein 4 |
| chr12_-_39734783 | 0.21 |
ENST00000552961.1
|
KIF21A
|
kinesin family member 21A |
| chr5_+_140254884 | 0.21 |
ENST00000398631.2
|
PCDHA12
|
protocadherin alpha 12 |
| chr4_-_152149033 | 0.21 |
ENST00000514152.1
|
SH3D19
|
SH3 domain containing 19 |
| chr4_-_85654615 | 0.21 |
ENST00000514711.1
|
WDFY3
|
WD repeat and FYVE domain containing 3 |
| chr3_-_151160938 | 0.20 |
ENST00000489791.1
|
IGSF10
|
immunoglobulin superfamily, member 10 |
| chr8_+_66619277 | 0.20 |
ENST00000521247.2
ENST00000527155.1 |
MTFR1
|
mitochondrial fission regulator 1 |
| chr4_-_83765613 | 0.20 |
ENST00000503937.1
|
SEC31A
|
SEC31 homolog A (S. cerevisiae) |
| chr5_-_177659625 | 0.20 |
ENST00000323594.4
|
PHYKPL
|
5-phosphohydroxy-L-lysine phospho-lyase |
| chr14_-_54955721 | 0.19 |
ENST00000554908.1
|
GMFB
|
glia maturation factor, beta |
| chr12_-_90049828 | 0.19 |
ENST00000261173.2
ENST00000348959.3 |
ATP2B1
|
ATPase, Ca++ transporting, plasma membrane 1 |
| chr17_+_28705921 | 0.19 |
ENST00000225719.4
|
CPD
|
carboxypeptidase D |
| chr4_+_169418195 | 0.19 |
ENST00000261509.6
ENST00000335742.7 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr17_+_40996590 | 0.19 |
ENST00000253799.3
ENST00000452774.2 |
AOC2
|
amine oxidase, copper containing 2 (retina-specific) |
| chr13_-_73301819 | 0.19 |
ENST00000377818.3
|
MZT1
|
mitotic spindle organizing protein 1 |
| chr7_-_55620433 | 0.18 |
ENST00000418904.1
|
VOPP1
|
vesicular, overexpressed in cancer, prosurvival protein 1 |
| chr4_-_71532339 | 0.18 |
ENST00000254801.4
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr10_-_63995871 | 0.18 |
ENST00000315289.2
|
RTKN2
|
rhotekin 2 |
| chr4_+_175205038 | 0.18 |
ENST00000457424.2
ENST00000514712.1 |
CEP44
|
centrosomal protein 44kDa |
| chr5_-_111093759 | 0.18 |
ENST00000509979.1
ENST00000513100.1 ENST00000508161.1 ENST00000455559.2 |
NREP
|
neuronal regeneration related protein |
| chr6_-_47009996 | 0.18 |
ENST00000371243.2
|
GPR110
|
G protein-coupled receptor 110 |
| chrX_+_37639264 | 0.18 |
ENST00000378588.4
|
CYBB
|
cytochrome b-245, beta polypeptide |
| chr12_+_94656297 | 0.18 |
ENST00000545312.1
|
PLXNC1
|
plexin C1 |
| chr16_-_1275257 | 0.18 |
ENST00000234798.4
|
TPSG1
|
tryptase gamma 1 |
| chr5_-_96518907 | 0.18 |
ENST00000508447.1
ENST00000283109.3 |
RIOK2
|
RIO kinase 2 |
| chr11_-_4629388 | 0.17 |
ENST00000526337.1
ENST00000300747.5 |
TRIM68
|
tripartite motif containing 68 |
| chr2_-_37544209 | 0.17 |
ENST00000234179.2
|
PRKD3
|
protein kinase D3 |
| chr17_+_35851570 | 0.17 |
ENST00000394386.1
|
DUSP14
|
dual specificity phosphatase 14 |
| chr5_+_35856951 | 0.17 |
ENST00000303115.3
ENST00000343305.4 ENST00000506850.1 ENST00000511982.1 |
IL7R
|
interleukin 7 receptor |
| chr11_+_112046190 | 0.17 |
ENST00000357685.5
ENST00000393032.2 ENST00000361053.4 |
BCO2
|
beta-carotene oxygenase 2 |
| chr12_-_28124903 | 0.17 |
ENST00000395872.1
ENST00000354417.3 ENST00000201015.4 |
PTHLH
|
parathyroid hormone-like hormone |
| chr1_+_196743943 | 0.16 |
ENST00000471440.2
ENST00000391985.3 |
CFHR3
|
complement factor H-related 3 |
| chr17_-_39023462 | 0.16 |
ENST00000251643.4
|
KRT12
|
keratin 12 |
| chr5_-_150138061 | 0.16 |
ENST00000521533.1
ENST00000424236.1 |
DCTN4
|
dynactin 4 (p62) |
| chr1_+_81771806 | 0.16 |
ENST00000370721.1
ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2
|
latrophilin 2 |
| chr16_+_53241854 | 0.16 |
ENST00000565803.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr14_+_77244349 | 0.16 |
ENST00000554743.1
|
VASH1
|
vasohibin 1 |
| chr7_+_79765071 | 0.16 |
ENST00000457358.2
|
GNAI1
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
| chr8_-_79717750 | 0.15 |
ENST00000263851.4
ENST00000379113.2 |
IL7
|
interleukin 7 |
| chr9_-_86593238 | 0.15 |
ENST00000351839.3
|
HNRNPK
|
heterogeneous nuclear ribonucleoprotein K |
| chr17_-_39165366 | 0.15 |
ENST00000391588.1
|
KRTAP3-1
|
keratin associated protein 3-1 |
| chr10_+_57358750 | 0.15 |
ENST00000512524.2
|
MTRNR2L5
|
MT-RNR2-like 5 |
| chr4_+_124571409 | 0.15 |
ENST00000514823.1
ENST00000511919.1 ENST00000508111.1 |
RP11-93L9.1
|
long intergenic non-protein coding RNA 1091 |
| chr5_+_140165876 | 0.15 |
ENST00000504120.2
ENST00000394633.3 ENST00000378133.3 |
PCDHA1
|
protocadherin alpha 1 |
| chr10_-_43892668 | 0.15 |
ENST00000544000.1
|
HNRNPF
|
heterogeneous nuclear ribonucleoprotein F |
| chr2_-_152589670 | 0.15 |
ENST00000604864.1
ENST00000603639.1 |
NEB
|
nebulin |
| chr8_-_101730061 | 0.15 |
ENST00000519100.1
|
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr4_-_100065440 | 0.15 |
ENST00000508393.1
ENST00000265512.7 |
ADH4
|
alcohol dehydrogenase 4 (class II), pi polypeptide |
| chr14_+_53196872 | 0.14 |
ENST00000442123.2
ENST00000354586.4 |
STYX
|
serine/threonine/tyrosine interacting protein |
| chr14_+_24540154 | 0.14 |
ENST00000559778.1
ENST00000560761.1 ENST00000557889.1 |
CPNE6
|
copine VI (neuronal) |
| chr8_+_21823726 | 0.14 |
ENST00000433566.4
|
XPO7
|
exportin 7 |
| chr7_+_75024903 | 0.14 |
ENST00000323819.3
ENST00000430211.1 |
TRIM73
|
tripartite motif containing 73 |
| chr1_-_160231451 | 0.14 |
ENST00000495887.1
|
DCAF8
|
DDB1 and CUL4 associated factor 8 |
| chr14_+_102276132 | 0.14 |
ENST00000350249.3
ENST00000557621.1 ENST00000556946.1 |
PPP2R5C
|
protein phosphatase 2, regulatory subunit B', gamma |
| chr7_-_130353553 | 0.14 |
ENST00000330992.7
ENST00000445977.2 |
COPG2
|
coatomer protein complex, subunit gamma 2 |
| chr1_+_196743912 | 0.14 |
ENST00000367425.4
|
CFHR3
|
complement factor H-related 3 |
| chr5_+_68530697 | 0.14 |
ENST00000256443.3
ENST00000514676.1 |
CDK7
|
cyclin-dependent kinase 7 |
| chr6_+_143999072 | 0.14 |
ENST00000440869.2
ENST00000367582.3 ENST00000451827.2 |
PHACTR2
|
phosphatase and actin regulator 2 |
| chr1_+_104159999 | 0.13 |
ENST00000414303.2
ENST00000423678.1 |
AMY2A
|
amylase, alpha 2A (pancreatic) |
| chr11_-_77122928 | 0.13 |
ENST00000528203.1
ENST00000528592.1 ENST00000528633.1 ENST00000529248.1 |
PAK1
|
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr18_-_57027194 | 0.13 |
ENST00000251047.5
|
LMAN1
|
lectin, mannose-binding, 1 |
| chr12_-_57328187 | 0.13 |
ENST00000293502.1
|
SDR9C7
|
short chain dehydrogenase/reductase family 9C, member 7 |
| chr7_+_141490017 | 0.13 |
ENST00000247883.4
|
TAS2R5
|
taste receptor, type 2, member 5 |
| chr14_+_67831576 | 0.13 |
ENST00000555876.1
|
EIF2S1
|
eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa |
| chr2_+_172309634 | 0.13 |
ENST00000339506.3
|
DCAF17
|
DDB1 and CUL4 associated factor 17 |
| chr4_+_156824840 | 0.13 |
ENST00000536354.2
|
TDO2
|
tryptophan 2,3-dioxygenase |
Network of associatons between targets according to the STRING database.
First level regulatory network of CDX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0002590 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 0.3 | 1.4 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.2 | 0.9 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 0.2 | 0.5 | GO:0060467 | negative regulation of fertilization(GO:0060467) negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.2 | 0.5 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 1.1 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 0.4 | GO:0046356 | acetyl-CoA catabolic process(GO:0046356) |
| 0.1 | 0.9 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.1 | 0.4 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.1 | 0.7 | GO:1903899 | lung goblet cell differentiation(GO:0060480) positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.1 | 0.5 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.1 | 0.3 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.1 | 0.6 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.6 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.1 | 0.3 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 0.1 | 0.9 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.1 | 0.3 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.1 | 0.2 | GO:0006425 | glutaminyl-tRNA aminoacylation(GO:0006425) |
| 0.1 | 0.3 | GO:0009446 | putrescine biosynthetic process(GO:0009446) |
| 0.1 | 0.7 | GO:0018406 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.1 | 0.3 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.1 | 0.3 | GO:1903615 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.1 | 0.1 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.1 | 0.1 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.1 | 0.3 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 0.4 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.1 | 0.3 | GO:1900133 | regulation of renin secretion into blood stream(GO:1900133) |
| 0.1 | 0.3 | GO:0044855 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.1 | 0.5 | GO:1990034 | cellular response to corticosterone stimulus(GO:0071386) calcium ion export from cell(GO:1990034) |
| 0.1 | 0.2 | GO:0071684 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.2 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.1 | GO:0006267 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.2 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.2 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.0 | 0.8 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.1 | GO:0001897 | cytolysis by symbiont of host cells(GO:0001897) |
| 0.0 | 0.7 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.0 | 0.2 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.0 | 0.3 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.3 | GO:0044789 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.0 | 0.1 | GO:1904640 | response to methionine(GO:1904640) |
| 0.0 | 0.2 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.0 | 0.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.2 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.2 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.4 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.1 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.6 | GO:1902373 | negative regulation of mRNA catabolic process(GO:1902373) |
| 0.0 | 0.3 | GO:0097338 | response to clozapine(GO:0097338) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.2 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.0 | 0.3 | GO:0042262 | DNA protection(GO:0042262) |
| 0.0 | 0.2 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.1 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.0 | 0.8 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.3 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.3 | GO:0045876 | positive regulation of sister chromatid cohesion(GO:0045876) |
| 0.0 | 0.2 | GO:0051418 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.1 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.4 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 0.1 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.0 | 1.8 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.4 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.1 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.0 | 0.2 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.3 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.9 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 | 0.3 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.2 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 2.0 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.0 | 0.1 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.5 | GO:0006293 | nucleotide-excision repair, preincision complex stabilization(GO:0006293) |
| 0.0 | 0.2 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.0 | 0.2 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.5 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.2 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.3 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.1 | GO:1902963 | regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) |
| 0.0 | 0.1 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.0 | 0.2 | GO:1904322 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 0.2 | GO:0006069 | ethanol oxidation(GO:0006069) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 0.9 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 1.9 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.1 | 0.2 | GO:0097635 | extrinsic component of autophagosome membrane(GO:0097635) |
| 0.1 | 0.5 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.2 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 0.1 | 0.5 | GO:0000798 | nuclear cohesin complex(GO:0000798) |
| 0.1 | 0.2 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 0.1 | GO:0043614 | multi-eIF complex(GO:0043614) translation preinitiation complex(GO:0070993) astrocyte end-foot(GO:0097450) |
| 0.0 | 0.1 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.0 | 0.9 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.1 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.0 | 0.1 | GO:0070985 | TFIIK complex(GO:0070985) |
| 0.0 | 1.1 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 0.5 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.3 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.0 | 0.3 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.4 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.0 | 0.5 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.2 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.1 | GO:0030430 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.0 | 0.2 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.0 | 0.1 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 0.3 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.1 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.9 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.3 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.2 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0015068 | amidinotransferase activity(GO:0015067) glycine amidinotransferase activity(GO:0015068) |
| 0.2 | 0.5 | GO:0015322 | proton-dependent oligopeptide secondary active transmembrane transporter activity(GO:0005427) secondary active oligopeptide transmembrane transporter activity(GO:0015322) |
| 0.2 | 0.9 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.1 | 1.1 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 0.9 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.1 | 0.4 | GO:0052594 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.1 | 0.3 | GO:0017130 | poly(C) RNA binding(GO:0017130) |
| 0.1 | 0.2 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.1 | 0.3 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.1 | 0.2 | GO:0060001 | minus-end directed microfilament motor activity(GO:0060001) |
| 0.1 | 0.4 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.1 | 0.2 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 0.1 | 1.0 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.1 | 0.7 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 0.8 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 0.3 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.1 | 0.7 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.1 | 0.2 | GO:0019115 | benzaldehyde dehydrogenase activity(GO:0019115) |
| 0.1 | 0.3 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.1 | 0.4 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.0 | 0.2 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.0 | 0.3 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.2 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.2 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.0 | 1.1 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.2 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 0.3 | GO:0030614 | oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) |
| 0.0 | 0.5 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.3 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.1 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.0 | 0.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.4 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.1 | GO:0004611 | phosphoenolpyruvate carboxykinase activity(GO:0004611) phosphoenolpyruvate carboxykinase (GTP) activity(GO:0004613) |
| 0.0 | 0.4 | GO:0050308 | carbohydrate phosphatase activity(GO:0019203) sugar-phosphatase activity(GO:0050308) |
| 0.0 | 0.3 | GO:0045029 | uridine nucleotide receptor activity(GO:0015065) UDP-activated nucleotide receptor activity(GO:0045029) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.1 | GO:1904854 | proteasome core complex binding(GO:1904854) |
| 0.0 | 0.1 | GO:0051499 | D-aminoacyl-tRNA deacylase activity(GO:0051499) D-tyrosyl-tRNA(Tyr) deacylase activity(GO:0051500) |
| 0.0 | 0.3 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.7 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.7 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.0 | 0.1 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.0 | 0.3 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.3 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.4 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 1.2 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.1 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.0 | 0.5 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.2 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.8 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.2 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.1 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 1.5 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.4 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 0.8 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 1.0 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 1.2 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 1.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.5 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 1.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 1.0 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 2.5 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.2 | REACTOME CLASS I MHC MEDIATED ANTIGEN PROCESSING PRESENTATION | Genes involved in Class I MHC mediated antigen processing & presentation |
| 0.0 | 0.6 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.5 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.3 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.4 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.3 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.1 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |