Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
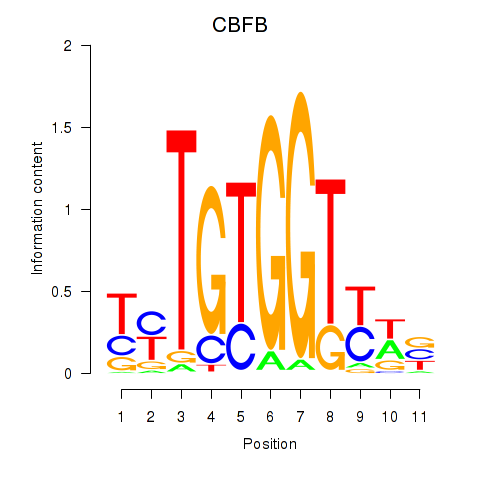
Results for CBFB
Z-value: 0.82
Transcription factors associated with CBFB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
CBFB
|
ENSG00000067955.9 | core-binding factor subunit beta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| CBFB | hg19_v2_chr16_+_67063036_67063088 | -0.98 | 2.2e-02 | Click! |
Activity profile of CBFB motif
Sorted Z-values of CBFB motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_34417479 | 2.54 |
ENST00000225245.5
|
CCL3
|
chemokine (C-C motif) ligand 3 |
| chr1_+_209859510 | 0.84 |
ENST00000367028.2
ENST00000261465.1 |
HSD11B1
|
hydroxysteroid (11-beta) dehydrogenase 1 |
| chr20_+_43343886 | 0.61 |
ENST00000190983.4
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr12_-_54778444 | 0.50 |
ENST00000551771.1
|
ZNF385A
|
zinc finger protein 385A |
| chr17_-_39183452 | 0.49 |
ENST00000361883.5
|
KRTAP1-5
|
keratin associated protein 1-5 |
| chr12_-_54778471 | 0.48 |
ENST00000550120.1
ENST00000394313.2 ENST00000547210.1 |
ZNF385A
|
zinc finger protein 385A |
| chr14_-_61116168 | 0.43 |
ENST00000247182.6
|
SIX1
|
SIX homeobox 1 |
| chr2_-_127977654 | 0.42 |
ENST00000409327.1
|
CYP27C1
|
cytochrome P450, family 27, subfamily C, polypeptide 1 |
| chr17_+_32582293 | 0.40 |
ENST00000580907.1
ENST00000225831.4 |
CCL2
|
chemokine (C-C motif) ligand 2 |
| chr11_+_114310164 | 0.39 |
ENST00000544196.1
ENST00000539754.1 ENST00000539275.1 |
REXO2
|
RNA exonuclease 2 |
| chr15_+_67390920 | 0.38 |
ENST00000559092.1
ENST00000560175.1 |
SMAD3
|
SMAD family member 3 |
| chrX_+_115567767 | 0.38 |
ENST00000371900.4
|
SLC6A14
|
solute carrier family 6 (amino acid transporter), member 14 |
| chrX_+_68048803 | 0.35 |
ENST00000204961.4
|
EFNB1
|
ephrin-B1 |
| chr11_+_114310102 | 0.32 |
ENST00000265881.5
|
REXO2
|
RNA exonuclease 2 |
| chr4_+_71091786 | 0.31 |
ENST00000317987.5
|
FDCSP
|
follicular dendritic cell secreted protein |
| chr14_-_59951112 | 0.29 |
ENST00000247194.4
|
L3HYPDH
|
L-3-hydroxyproline dehydratase (trans-) |
| chr4_+_74347400 | 0.28 |
ENST00000226355.3
|
AFM
|
afamin |
| chr9_-_130541017 | 0.28 |
ENST00000314830.8
|
SH2D3C
|
SH2 domain containing 3C |
| chr1_+_66796401 | 0.26 |
ENST00000528771.1
|
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr11_-_64885111 | 0.26 |
ENST00000528598.1
ENST00000310597.4 |
ZNHIT2
|
zinc finger, HIT-type containing 2 |
| chrX_-_49041242 | 0.25 |
ENST00000453382.1
ENST00000540849.1 ENST00000536904.1 ENST00000432913.1 |
PRICKLE3
|
prickle homolog 3 (Drosophila) |
| chr2_+_234826016 | 0.24 |
ENST00000324695.4
ENST00000433712.2 |
TRPM8
|
transient receptor potential cation channel, subfamily M, member 8 |
| chr1_-_179112173 | 0.23 |
ENST00000408940.3
ENST00000504405.1 |
ABL2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr11_-_66103867 | 0.22 |
ENST00000424433.2
|
RIN1
|
Ras and Rab interactor 1 |
| chr11_-_66103932 | 0.22 |
ENST00000311320.4
|
RIN1
|
Ras and Rab interactor 1 |
| chrX_-_19688475 | 0.21 |
ENST00000541422.1
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr12_-_54778244 | 0.21 |
ENST00000549937.1
|
ZNF385A
|
zinc finger protein 385A |
| chr5_+_131409476 | 0.20 |
ENST00000296871.2
|
CSF2
|
colony stimulating factor 2 (granulocyte-macrophage) |
| chr13_+_76210448 | 0.19 |
ENST00000377499.5
|
LMO7
|
LIM domain 7 |
| chr5_-_39203093 | 0.19 |
ENST00000515010.1
|
FYB
|
FYN binding protein |
| chr11_-_66104237 | 0.16 |
ENST00000530056.1
|
RIN1
|
Ras and Rab interactor 1 |
| chr1_-_179112189 | 0.16 |
ENST00000512653.1
ENST00000344730.3 |
ABL2
|
c-abl oncogene 2, non-receptor tyrosine kinase |
| chr6_+_30908747 | 0.15 |
ENST00000462446.1
ENST00000304311.2 |
DPCR1
|
diffuse panbronchiolitis critical region 1 |
| chr8_+_28747884 | 0.14 |
ENST00000287701.10
ENST00000444075.1 ENST00000403668.2 ENST00000519662.1 ENST00000558662.1 ENST00000523613.1 ENST00000560599.1 ENST00000397358.3 |
HMBOX1
|
homeobox containing 1 |
| chr3_+_98482159 | 0.14 |
ENST00000468553.1
|
ST3GAL6
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chr6_+_45296391 | 0.13 |
ENST00000371436.6
ENST00000576263.1 |
RUNX2
|
runt-related transcription factor 2 |
| chr6_-_152623231 | 0.13 |
ENST00000540663.1
ENST00000537033.1 |
SYNE1
|
spectrin repeat containing, nuclear envelope 1 |
| chr3_+_156799587 | 0.12 |
ENST00000469196.1
|
RP11-6F2.5
|
RP11-6F2.5 |
| chr2_-_87248975 | 0.12 |
ENST00000409310.2
ENST00000355705.3 |
PLGLB1
|
plasminogen-like B1 |
| chr3_-_99569821 | 0.12 |
ENST00000487087.1
|
FILIP1L
|
filamin A interacting protein 1-like |
| chr19_-_14048804 | 0.11 |
ENST00000254320.3
ENST00000586075.1 |
PODNL1
|
podocan-like 1 |
| chr12_-_4754339 | 0.11 |
ENST00000228850.1
|
AKAP3
|
A kinase (PRKA) anchor protein 3 |
| chr1_+_89246647 | 0.10 |
ENST00000544045.1
|
PKN2
|
protein kinase N2 |
| chr4_+_88896819 | 0.10 |
ENST00000237623.7
ENST00000395080.3 ENST00000508233.1 ENST00000360804.4 |
SPP1
|
secreted phosphoprotein 1 |
| chr15_-_64385981 | 0.10 |
ENST00000557835.1
ENST00000380290.3 ENST00000559950.1 |
FAM96A
|
family with sequence similarity 96, member A |
| chr2_-_158300556 | 0.10 |
ENST00000264192.3
|
CYTIP
|
cytohesin 1 interacting protein |
| chr3_+_98482175 | 0.09 |
ENST00000485391.1
ENST00000492254.1 |
ST3GAL6
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chr1_-_157014865 | 0.08 |
ENST00000361409.2
|
ARHGEF11
|
Rho guanine nucleotide exchange factor (GEF) 11 |
| chr2_+_68592305 | 0.08 |
ENST00000234313.7
|
PLEK
|
pleckstrin |
| chr8_-_134072593 | 0.07 |
ENST00000427060.2
|
SLA
|
Src-like-adaptor |
| chr8_-_60031762 | 0.07 |
ENST00000361421.1
|
TOX
|
thymocyte selection-associated high mobility group box |
| chr3_+_157154578 | 0.06 |
ENST00000295927.3
|
PTX3
|
pentraxin 3, long |
| chr12_-_14849470 | 0.06 |
ENST00000261170.3
|
GUCY2C
|
guanylate cyclase 2C (heat stable enterotoxin receptor) |
| chr15_-_64386120 | 0.06 |
ENST00000300030.3
|
FAM96A
|
family with sequence similarity 96, member A |
| chr2_+_103035102 | 0.06 |
ENST00000264260.2
|
IL18RAP
|
interleukin 18 receptor accessory protein |
| chr1_+_47264711 | 0.06 |
ENST00000371923.4
ENST00000271153.4 ENST00000371919.4 |
CYP4B1
|
cytochrome P450, family 4, subfamily B, polypeptide 1 |
| chr2_+_207024306 | 0.06 |
ENST00000236957.5
ENST00000392221.1 ENST00000392222.2 ENST00000445505.1 |
EEF1B2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr11_+_114310237 | 0.05 |
ENST00000539119.1
|
REXO2
|
RNA exonuclease 2 |
| chr19_-_14049184 | 0.05 |
ENST00000339560.5
|
PODNL1
|
podocan-like 1 |
| chr8_-_28747717 | 0.05 |
ENST00000416984.2
|
INTS9
|
integrator complex subunit 9 |
| chr15_+_64386261 | 0.04 |
ENST00000560829.1
|
SNX1
|
sorting nexin 1 |
| chr20_+_54987168 | 0.04 |
ENST00000360314.3
|
CASS4
|
Cas scaffolding protein family member 4 |
| chr9_+_123884038 | 0.04 |
ENST00000373847.1
|
CNTRL
|
centriolin |
| chr14_+_72064945 | 0.04 |
ENST00000537413.1
|
SIPA1L1
|
signal-induced proliferation-associated 1 like 1 |
| chr15_-_64665911 | 0.04 |
ENST00000606793.1
ENST00000561349.1 ENST00000560278.1 |
CTD-2116N17.1
|
Uncharacterized protein |
| chr9_-_123342415 | 0.03 |
ENST00000349780.4
ENST00000360190.4 ENST00000360822.3 ENST00000359309.3 |
CDK5RAP2
|
CDK5 regulatory subunit associated protein 2 |
| chr8_-_28747424 | 0.03 |
ENST00000523436.1
ENST00000397363.4 ENST00000521777.1 ENST00000520184.1 ENST00000521022.1 |
INTS9
|
integrator complex subunit 9 |
| chr19_+_10541462 | 0.03 |
ENST00000293683.5
|
PDE4A
|
phosphodiesterase 4A, cAMP-specific |
| chr2_+_173955327 | 0.03 |
ENST00000422149.1
|
MLTK
|
Mitogen-activated protein kinase kinase kinase MLT |
| chr5_-_137878887 | 0.03 |
ENST00000507939.1
ENST00000572514.1 ENST00000499810.2 ENST00000360541.5 |
ETF1
|
eukaryotic translation termination factor 1 |
| chr14_+_85996507 | 0.03 |
ENST00000554746.1
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2 |
| chr2_-_207024233 | 0.02 |
ENST00000423725.1
ENST00000233190.6 |
NDUFS1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr5_+_67576109 | 0.02 |
ENST00000522084.1
|
PIK3R1
|
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr1_+_159770292 | 0.02 |
ENST00000536257.1
ENST00000321935.6 |
FCRL6
|
Fc receptor-like 6 |
| chr8_+_67624653 | 0.02 |
ENST00000521198.2
|
SGK3
|
serum/glucocorticoid regulated kinase family, member 3 |
| chrX_-_70838306 | 0.01 |
ENST00000373691.4
ENST00000373693.3 |
CXCR3
|
chemokine (C-X-C motif) receptor 3 |
| chr1_+_39876151 | 0.00 |
ENST00000530275.1
|
KIAA0754
|
KIAA0754 |
| chr12_-_52585765 | 0.00 |
ENST00000313234.5
ENST00000394815.2 |
KRT80
|
keratin 80 |
Network of associatons between targets according to the STRING database.
First level regulatory network of CBFB
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.5 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.2 | 1.2 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.1 | 0.4 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.1 | 0.4 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.1 | 0.6 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.1 | 0.2 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.1 | 0.4 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.8 | GO:0006704 | glucocorticoid biosynthetic process(GO:0006704) |
| 0.0 | 0.2 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.1 | GO:0035938 | estrogen secretion(GO:0035937) estradiol secretion(GO:0035938) regulation of estrogen secretion(GO:2000861) regulation of estradiol secretion(GO:2000864) |
| 0.0 | 0.3 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.4 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.1 | GO:0044793 | negative regulation by host of viral process(GO:0044793) |
| 0.0 | 0.8 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 | 0.1 | GO:0033625 | positive regulation of integrin activation(GO:0033625) |
| 0.0 | 0.1 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.1 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.5 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) CCR1 chemokine receptor binding(GO:0031726) |
| 0.3 | 0.8 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.1 | 0.4 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.1 | 0.3 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.1 | 0.2 | GO:0016855 | racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.1 | 0.4 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.2 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.0 | 0.4 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.4 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.6 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.8 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.4 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.0 | 0.1 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.6 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.4 | PID IL23 PATHWAY | IL23-mediated signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.0 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.8 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |