Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for CAGUGCA
Z-value: 0.67
miRNA associated with seed CAGUGCA
| Name | miRBASE accession |
|---|---|
|
hsa-miR-148a-3p
|
MIMAT0000243 |
|
hsa-miR-148b-3p
|
MIMAT0000759 |
|
hsa-miR-152-3p
|
MIMAT0000438 |
Activity profile of CAGUGCA motif
Sorted Z-values of CAGUGCA motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_182545603 | 0.46 |
ENST00000295108.3
|
NEUROD1
|
neuronal differentiation 1 |
| chr17_-_66287257 | 0.27 |
ENST00000327268.4
|
SLC16A6
|
solute carrier family 16, member 6 |
| chr7_+_100770328 | 0.26 |
ENST00000223095.4
ENST00000445463.2 |
SERPINE1
|
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
| chr6_+_144471643 | 0.26 |
ENST00000367568.4
|
STX11
|
syntaxin 11 |
| chr5_-_90679145 | 0.24 |
ENST00000265138.3
|
ARRDC3
|
arrestin domain containing 3 |
| chr8_-_74791051 | 0.24 |
ENST00000453587.2
ENST00000602969.1 ENST00000602593.1 ENST00000419880.3 ENST00000517608.1 |
UBE2W
|
ubiquitin-conjugating enzyme E2W (putative) |
| chr16_+_69599861 | 0.22 |
ENST00000354436.2
|
NFAT5
|
nuclear factor of activated T-cells 5, tonicity-responsive |
| chr6_+_17281573 | 0.21 |
ENST00000379052.5
|
RBM24
|
RNA binding motif protein 24 |
| chr5_+_122110691 | 0.21 |
ENST00000379516.2
ENST00000505934.1 ENST00000514949.1 |
SNX2
|
sorting nexin 2 |
| chr3_+_194406603 | 0.21 |
ENST00000329759.4
|
FAM43A
|
family with sequence similarity 43, member A |
| chr5_-_98262240 | 0.21 |
ENST00000284049.3
|
CHD1
|
chromodomain helicase DNA binding protein 1 |
| chr2_-_36825281 | 0.20 |
ENST00000405912.3
ENST00000379245.4 |
FEZ2
|
fasciculation and elongation protein zeta 2 (zygin II) |
| chr17_+_54671047 | 0.20 |
ENST00000332822.4
|
NOG
|
noggin |
| chr12_+_111843749 | 0.20 |
ENST00000341259.2
|
SH2B3
|
SH2B adaptor protein 3 |
| chr21_-_39288743 | 0.20 |
ENST00000609713.1
|
KCNJ6
|
potassium inwardly-rectifying channel, subfamily J, member 6 |
| chr3_-_100120223 | 0.20 |
ENST00000284320.5
|
TOMM70A
|
translocase of outer mitochondrial membrane 70 homolog A (S. cerevisiae) |
| chr5_-_39074479 | 0.20 |
ENST00000514735.1
ENST00000296782.5 ENST00000357387.3 |
RICTOR
|
RPTOR independent companion of MTOR, complex 2 |
| chr18_+_60190226 | 0.19 |
ENST00000269499.5
|
ZCCHC2
|
zinc finger, CCHC domain containing 2 |
| chr15_+_41221536 | 0.19 |
ENST00000249749.5
|
DLL4
|
delta-like 4 (Drosophila) |
| chr5_+_78532003 | 0.18 |
ENST00000396137.4
|
JMY
|
junction mediating and regulatory protein, p53 cofactor |
| chr11_-_59436453 | 0.17 |
ENST00000300146.9
|
PATL1
|
protein associated with topoisomerase II homolog 1 (yeast) |
| chr13_-_50510434 | 0.17 |
ENST00000361840.3
|
SPRYD7
|
SPRY domain containing 7 |
| chr7_+_116312411 | 0.16 |
ENST00000456159.1
ENST00000397752.3 ENST00000318493.6 |
MET
|
met proto-oncogene |
| chr1_-_51984908 | 0.16 |
ENST00000371730.2
|
EPS15
|
epidermal growth factor receptor pathway substrate 15 |
| chr2_+_5832799 | 0.15 |
ENST00000322002.3
|
SOX11
|
SRY (sex determining region Y)-box 11 |
| chr1_-_22109682 | 0.15 |
ENST00000400301.1
ENST00000532737.1 |
USP48
|
ubiquitin specific peptidase 48 |
| chr20_-_48330377 | 0.15 |
ENST00000371711.4
|
B4GALT5
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 |
| chr5_+_179125907 | 0.15 |
ENST00000247461.4
ENST00000452673.2 ENST00000502498.1 ENST00000507307.1 ENST00000513246.1 ENST00000502673.1 ENST00000506654.1 ENST00000512607.2 ENST00000510810.1 |
CANX
|
calnexin |
| chr12_-_26278030 | 0.14 |
ENST00000242728.4
|
BHLHE41
|
basic helix-loop-helix family, member e41 |
| chr14_-_50698276 | 0.14 |
ENST00000216373.5
|
SOS2
|
son of sevenless homolog 2 (Drosophila) |
| chr17_-_78450398 | 0.14 |
ENST00000306773.4
|
NPTX1
|
neuronal pentraxin I |
| chr5_+_96271141 | 0.14 |
ENST00000231368.5
|
LNPEP
|
leucyl/cystinyl aminopeptidase |
| chr13_-_79233314 | 0.14 |
ENST00000282003.6
|
RNF219
|
ring finger protein 219 |
| chrX_+_17393543 | 0.14 |
ENST00000380060.3
|
NHS
|
Nance-Horan syndrome (congenital cataracts and dental anomalies) |
| chr5_-_126366500 | 0.14 |
ENST00000308660.5
|
MARCH3
|
membrane-associated ring finger (C3HC4) 3, E3 ubiquitin protein ligase |
| chr20_-_5591626 | 0.13 |
ENST00000379019.4
|
GPCPD1
|
glycerophosphocholine phosphodiesterase GDE1 homolog (S. cerevisiae) |
| chr13_+_28813645 | 0.13 |
ENST00000282391.5
|
PAN3
|
PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr20_-_524455 | 0.13 |
ENST00000349736.5
ENST00000217244.3 |
CSNK2A1
|
casein kinase 2, alpha 1 polypeptide |
| chrX_-_134049262 | 0.13 |
ENST00000370783.3
|
MOSPD1
|
motile sperm domain containing 1 |
| chr1_+_70671363 | 0.13 |
ENST00000370951.1
ENST00000370950.3 ENST00000405432.1 ENST00000454435.2 |
SRSF11
|
serine/arginine-rich splicing factor 11 |
| chr10_+_24755416 | 0.12 |
ENST00000396446.1
ENST00000396445.1 ENST00000376451.2 |
KIAA1217
|
KIAA1217 |
| chr19_+_926000 | 0.12 |
ENST00000263620.3
|
ARID3A
|
AT rich interactive domain 3A (BRIGHT-like) |
| chr18_+_43753974 | 0.12 |
ENST00000282059.6
ENST00000321319.6 |
C18orf25
|
chromosome 18 open reading frame 25 |
| chr18_-_19180681 | 0.12 |
ENST00000269214.5
|
ESCO1
|
establishment of sister chromatid cohesion N-acetyltransferase 1 |
| chr1_+_162467595 | 0.12 |
ENST00000538489.1
ENST00000489294.1 |
UHMK1
|
U2AF homology motif (UHM) kinase 1 |
| chr3_-_172428959 | 0.11 |
ENST00000475381.1
ENST00000538775.1 ENST00000273512.3 ENST00000543711.1 |
NCEH1
|
neutral cholesterol ester hydrolase 1 |
| chr18_-_45456930 | 0.11 |
ENST00000262160.6
ENST00000587269.1 |
SMAD2
|
SMAD family member 2 |
| chr2_-_9143786 | 0.11 |
ENST00000462696.1
ENST00000305997.3 |
MBOAT2
|
membrane bound O-acyltransferase domain containing 2 |
| chrX_+_40440146 | 0.11 |
ENST00000535539.1
ENST00000378438.4 ENST00000436783.1 ENST00000544975.1 ENST00000535777.1 ENST00000447485.1 ENST00000423649.1 |
ATP6AP2
|
ATPase, H+ transporting, lysosomal accessory protein 2 |
| chr3_+_138066539 | 0.10 |
ENST00000289104.4
|
MRAS
|
muscle RAS oncogene homolog |
| chr5_-_78809950 | 0.10 |
ENST00000334082.6
|
HOMER1
|
homer homolog 1 (Drosophila) |
| chr4_-_103748880 | 0.10 |
ENST00000453744.2
ENST00000349311.8 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr5_+_78908233 | 0.10 |
ENST00000453514.1
ENST00000423041.2 ENST00000504233.1 ENST00000428308.2 |
PAPD4
|
PAP associated domain containing 4 |
| chr19_-_39805976 | 0.10 |
ENST00000248668.4
|
LRFN1
|
leucine rich repeat and fibronectin type III domain containing 1 |
| chr2_-_39348137 | 0.10 |
ENST00000426016.1
|
SOS1
|
son of sevenless homolog 1 (Drosophila) |
| chr5_+_34656331 | 0.10 |
ENST00000265109.3
|
RAI14
|
retinoic acid induced 14 |
| chr8_+_29952914 | 0.09 |
ENST00000321250.8
ENST00000518001.1 ENST00000520682.1 ENST00000442880.2 ENST00000523116.1 |
LEPROTL1
|
leptin receptor overlapping transcript-like 1 |
| chr1_+_24286287 | 0.09 |
ENST00000334351.7
ENST00000374468.1 |
PNRC2
|
proline-rich nuclear receptor coactivator 2 |
| chr1_+_204494618 | 0.09 |
ENST00000367180.1
ENST00000391947.2 |
MDM4
|
Mdm4 p53 binding protein homolog (mouse) |
| chr1_-_46598284 | 0.09 |
ENST00000423209.1
ENST00000262741.5 |
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
| chr11_-_118661828 | 0.09 |
ENST00000264018.4
|
DDX6
|
DEAD (Asp-Glu-Ala-Asp) box helicase 6 |
| chr2_-_42180940 | 0.09 |
ENST00000378711.2
|
C2orf91
|
chromosome 2 open reading frame 91 |
| chr2_+_70142189 | 0.09 |
ENST00000264444.2
|
MXD1
|
MAX dimerization protein 1 |
| chr17_+_57784826 | 0.09 |
ENST00000262291.4
|
VMP1
|
vacuole membrane protein 1 |
| chr17_-_53499310 | 0.09 |
ENST00000262065.3
|
MMD
|
monocyte to macrophage differentiation-associated |
| chr19_+_7598890 | 0.09 |
ENST00000221249.6
ENST00000601668.1 ENST00000601001.1 |
PNPLA6
|
patatin-like phospholipase domain containing 6 |
| chr12_+_79258547 | 0.09 |
ENST00000457153.2
|
SYT1
|
synaptotagmin I |
| chr11_-_115375107 | 0.09 |
ENST00000545380.1
ENST00000452722.3 ENST00000537058.1 ENST00000536727.1 ENST00000542447.2 ENST00000331581.6 |
CADM1
|
cell adhesion molecule 1 |
| chr15_-_52030293 | 0.09 |
ENST00000560491.1
ENST00000267838.3 |
LYSMD2
|
LysM, putative peptidoglycan-binding, domain containing 2 |
| chr3_+_113666748 | 0.09 |
ENST00000330212.3
ENST00000498275.1 |
ZDHHC23
|
zinc finger, DHHC-type containing 23 |
| chr5_-_59189545 | 0.09 |
ENST00000340635.6
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr2_-_180129484 | 0.09 |
ENST00000428443.3
|
SESTD1
|
SEC14 and spectrin domains 1 |
| chr17_+_47865917 | 0.08 |
ENST00000259021.4
ENST00000454930.2 ENST00000509773.1 ENST00000510819.1 ENST00000424009.2 |
KAT7
|
K(lysine) acetyltransferase 7 |
| chr8_+_11141925 | 0.08 |
ENST00000221086.3
|
MTMR9
|
myotubularin related protein 9 |
| chr15_-_41408409 | 0.08 |
ENST00000361937.3
|
INO80
|
INO80 complex subunit |
| chr3_-_197476560 | 0.08 |
ENST00000273582.5
|
KIAA0226
|
KIAA0226 |
| chr13_-_21476900 | 0.08 |
ENST00000400602.2
ENST00000255305.6 |
XPO4
|
exportin 4 |
| chr17_-_71228357 | 0.08 |
ENST00000583024.1
ENST00000403627.3 ENST00000405159.3 ENST00000581110.1 |
FAM104A
|
family with sequence similarity 104, member A |
| chr2_-_9695847 | 0.08 |
ENST00000310823.3
ENST00000497134.1 |
ADAM17
|
ADAM metallopeptidase domain 17 |
| chr11_-_45687128 | 0.08 |
ENST00000308064.2
|
CHST1
|
carbohydrate (keratan sulfate Gal-6) sulfotransferase 1 |
| chr18_+_8609402 | 0.08 |
ENST00000329286.6
|
RAB12
|
RAB12, member RAS oncogene family |
| chr12_-_54813229 | 0.08 |
ENST00000293379.4
|
ITGA5
|
integrin, alpha 5 (fibronectin receptor, alpha polypeptide) |
| chr6_-_170599561 | 0.08 |
ENST00000366756.3
|
DLL1
|
delta-like 1 (Drosophila) |
| chr3_-_79068594 | 0.08 |
ENST00000436010.2
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr15_+_44038590 | 0.07 |
ENST00000300289.5
ENST00000538521.1 |
PDIA3
|
protein disulfide isomerase family A, member 3 |
| chr11_+_109964087 | 0.07 |
ENST00000278590.3
|
ZC3H12C
|
zinc finger CCCH-type containing 12C |
| chr20_-_52210368 | 0.07 |
ENST00000371471.2
|
ZNF217
|
zinc finger protein 217 |
| chr17_-_62658186 | 0.07 |
ENST00000262435.9
|
SMURF2
|
SMAD specific E3 ubiquitin protein ligase 2 |
| chr4_+_52709229 | 0.07 |
ENST00000334635.5
ENST00000381441.3 ENST00000381437.4 |
DCUN1D4
|
DCN1, defective in cullin neddylation 1, domain containing 4 |
| chr10_-_33623564 | 0.07 |
ENST00000374875.1
ENST00000374822.4 |
NRP1
|
neuropilin 1 |
| chr5_+_102594403 | 0.07 |
ENST00000319933.2
|
C5orf30
|
chromosome 5 open reading frame 30 |
| chr21_-_43299404 | 0.07 |
ENST00000422911.1
ENST00000398548.1 ENST00000538201.1 ENST00000269844.3 |
PRDM15
|
PR domain containing 15 |
| chr20_+_43514315 | 0.07 |
ENST00000353703.4
|
YWHAB
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta |
| chr7_+_55086794 | 0.07 |
ENST00000275493.2
ENST00000442591.1 |
EGFR
|
epidermal growth factor receptor |
| chr4_+_128703295 | 0.07 |
ENST00000296464.4
ENST00000508549.1 |
HSPA4L
|
heat shock 70kDa protein 4-like |
| chr16_+_30935418 | 0.07 |
ENST00000338343.4
|
FBXL19
|
F-box and leucine-rich repeat protein 19 |
| chr22_+_18121562 | 0.07 |
ENST00000355028.3
|
BCL2L13
|
BCL2-like 13 (apoptosis facilitator) |
| chr7_+_116139424 | 0.07 |
ENST00000222693.4
|
CAV2
|
caveolin 2 |
| chr15_-_52861394 | 0.06 |
ENST00000563277.1
ENST00000566423.1 |
ARPP19
|
cAMP-regulated phosphoprotein, 19kDa |
| chr6_+_64345698 | 0.06 |
ENST00000506783.1
ENST00000481385.2 ENST00000515594.1 ENST00000494284.2 ENST00000262043.3 |
PHF3
|
PHD finger protein 3 |
| chr1_-_8086343 | 0.06 |
ENST00000474874.1
ENST00000469499.1 ENST00000377482.5 |
ERRFI1
|
ERBB receptor feedback inhibitor 1 |
| chr7_-_139876812 | 0.06 |
ENST00000397560.2
|
JHDM1D
|
lysine (K)-specific demethylase 7A |
| chr1_-_78225482 | 0.06 |
ENST00000524778.1
ENST00000370794.3 ENST00000370793.1 ENST00000370792.3 |
USP33
|
ubiquitin specific peptidase 33 |
| chr5_+_112849373 | 0.06 |
ENST00000161863.4
ENST00000515883.1 |
YTHDC2
|
YTH domain containing 2 |
| chr2_+_46524537 | 0.06 |
ENST00000263734.3
|
EPAS1
|
endothelial PAS domain protein 1 |
| chr12_-_8025442 | 0.06 |
ENST00000340749.5
ENST00000535295.1 ENST00000539234.1 |
SLC2A14
|
solute carrier family 2 (facilitated glucose transporter), member 14 |
| chr18_+_59854480 | 0.06 |
ENST00000256858.6
ENST00000398130.2 |
KIAA1468
|
KIAA1468 |
| chr19_-_46476791 | 0.06 |
ENST00000263257.5
|
NOVA2
|
neuro-oncological ventral antigen 2 |
| chr2_-_70780770 | 0.06 |
ENST00000444975.1
ENST00000445399.1 ENST00000418333.2 |
TGFA
|
transforming growth factor, alpha |
| chr7_-_123389104 | 0.06 |
ENST00000223023.4
|
WASL
|
Wiskott-Aldrich syndrome-like |
| chr6_+_15246501 | 0.06 |
ENST00000341776.2
|
JARID2
|
jumonji, AT rich interactive domain 2 |
| chr1_-_109584608 | 0.06 |
ENST00000400794.3
ENST00000528747.1 ENST00000369962.3 ENST00000361054.3 |
WDR47
|
WD repeat domain 47 |
| chr9_-_140196703 | 0.06 |
ENST00000356628.2
|
NRARP
|
NOTCH-regulated ankyrin repeat protein |
| chr5_+_179921430 | 0.06 |
ENST00000393356.1
|
CNOT6
|
CCR4-NOT transcription complex, subunit 6 |
| chr2_+_228336849 | 0.06 |
ENST00000409979.2
ENST00000310078.8 |
AGFG1
|
ArfGAP with FG repeats 1 |
| chr1_-_27930102 | 0.06 |
ENST00000247087.5
ENST00000374011.2 |
AHDC1
|
AT hook, DNA binding motif, containing 1 |
| chr17_+_16593539 | 0.06 |
ENST00000340621.5
ENST00000399273.1 ENST00000443444.2 ENST00000360524.8 ENST00000456009.1 |
CCDC144A
|
coiled-coil domain containing 144A |
| chr14_-_91526922 | 0.06 |
ENST00000418736.2
ENST00000261991.3 |
RPS6KA5
|
ribosomal protein S6 kinase, 90kDa, polypeptide 5 |
| chr17_+_28705921 | 0.06 |
ENST00000225719.4
|
CPD
|
carboxypeptidase D |
| chr18_-_57027194 | 0.05 |
ENST00000251047.5
|
LMAN1
|
lectin, mannose-binding, 1 |
| chr1_-_157108130 | 0.05 |
ENST00000368192.4
|
ETV3
|
ets variant 3 |
| chr3_+_178866199 | 0.05 |
ENST00000263967.3
|
PIK3CA
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha |
| chr12_+_104324112 | 0.05 |
ENST00000299767.5
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1 |
| chrX_+_16804544 | 0.05 |
ENST00000380122.5
ENST00000398155.4 |
TXLNG
|
taxilin gamma |
| chr22_+_30115986 | 0.05 |
ENST00000216144.3
|
CABP7
|
calcium binding protein 7 |
| chr11_+_130318869 | 0.05 |
ENST00000299164.2
|
ADAMTS15
|
ADAM metallopeptidase with thrombospondin type 1 motif, 15 |
| chr21_-_34100244 | 0.05 |
ENST00000382491.3
ENST00000357345.3 ENST00000429236.1 |
SYNJ1
|
synaptojanin 1 |
| chr2_-_183903133 | 0.05 |
ENST00000361354.4
|
NCKAP1
|
NCK-associated protein 1 |
| chr1_-_173991434 | 0.05 |
ENST00000367696.2
|
RC3H1
|
ring finger and CCCH-type domains 1 |
| chr9_+_91003271 | 0.05 |
ENST00000375859.3
ENST00000541629.1 |
SPIN1
|
spindlin 1 |
| chr7_-_105517021 | 0.05 |
ENST00000318724.4
ENST00000419735.3 |
ATXN7L1
|
ataxin 7-like 1 |
| chr1_+_36348790 | 0.05 |
ENST00000373204.4
|
AGO1
|
argonaute RISC catalytic component 1 |
| chr3_+_5163905 | 0.05 |
ENST00000256496.3
ENST00000419534.2 |
ARL8B
|
ADP-ribosylation factor-like 8B |
| chr1_-_35325400 | 0.05 |
ENST00000521580.2
|
SMIM12
|
small integral membrane protein 12 |
| chr2_+_86947296 | 0.05 |
ENST00000283632.4
|
RMND5A
|
required for meiotic nuclear division 5 homolog A (S. cerevisiae) |
| chr17_-_1532106 | 0.05 |
ENST00000301335.5
ENST00000382147.4 |
SLC43A2
|
solute carrier family 43 (amino acid system L transporter), member 2 |
| chr16_-_71843047 | 0.04 |
ENST00000299980.4
ENST00000393512.3 |
AP1G1
|
adaptor-related protein complex 1, gamma 1 subunit |
| chr15_-_48937982 | 0.04 |
ENST00000316623.5
|
FBN1
|
fibrillin 1 |
| chr17_-_37607497 | 0.04 |
ENST00000394287.3
ENST00000300651.6 |
MED1
|
mediator complex subunit 1 |
| chr12_-_50561075 | 0.04 |
ENST00000422340.2
ENST00000317551.6 |
CERS5
|
ceramide synthase 5 |
| chr1_-_161102421 | 0.04 |
ENST00000490843.2
ENST00000368006.3 ENST00000392188.1 ENST00000545495.1 |
DEDD
|
death effector domain containing |
| chr1_+_26737253 | 0.04 |
ENST00000326279.6
|
LIN28A
|
lin-28 homolog A (C. elegans) |
| chr16_-_73082274 | 0.04 |
ENST00000268489.5
|
ZFHX3
|
zinc finger homeobox 3 |
| chrX_-_19905703 | 0.04 |
ENST00000397821.3
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chrX_+_146993449 | 0.04 |
ENST00000218200.8
ENST00000370471.3 ENST00000370477.1 |
FMR1
|
fragile X mental retardation 1 |
| chr1_-_153895377 | 0.04 |
ENST00000368655.4
|
GATAD2B
|
GATA zinc finger domain containing 2B |
| chr8_+_35649365 | 0.04 |
ENST00000437887.1
|
AC012215.1
|
Uncharacterized protein |
| chr3_+_9691117 | 0.04 |
ENST00000353332.5
ENST00000420925.1 ENST00000296003.4 ENST00000351233.5 |
MTMR14
|
myotubularin related protein 14 |
| chr2_+_24714729 | 0.04 |
ENST00000406961.1
ENST00000405141.1 |
NCOA1
|
nuclear receptor coactivator 1 |
| chr2_-_190649082 | 0.04 |
ENST00000392350.3
ENST00000392349.4 |
ORMDL1
|
ORM1-like 1 (S. cerevisiae) |
| chr3_+_38495333 | 0.04 |
ENST00000352511.4
|
ACVR2B
|
activin A receptor, type IIB |
| chr16_-_69419871 | 0.04 |
ENST00000603068.1
ENST00000254942.3 ENST00000567296.2 |
TERF2
|
telomeric repeat binding factor 2 |
| chrX_-_24690771 | 0.04 |
ENST00000379145.1
|
PCYT1B
|
phosphate cytidylyltransferase 1, choline, beta |
| chr20_-_31071239 | 0.04 |
ENST00000359676.5
|
C20orf112
|
chromosome 20 open reading frame 112 |
| chr3_-_64211112 | 0.04 |
ENST00000295902.6
|
PRICKLE2
|
prickle homolog 2 (Drosophila) |
| chr10_+_93683519 | 0.04 |
ENST00000265990.6
|
BTAF1
|
BTAF1 RNA polymerase II, B-TFIID transcription factor-associated, 170kDa |
| chr4_+_39046615 | 0.04 |
ENST00000261425.3
ENST00000508137.2 |
KLHL5
|
kelch-like family member 5 |
| chr10_+_60272814 | 0.04 |
ENST00000373886.3
|
BICC1
|
bicaudal C homolog 1 (Drosophila) |
| chr5_+_118407053 | 0.04 |
ENST00000311085.8
ENST00000539542.1 |
DMXL1
|
Dmx-like 1 |
| chr14_-_92572894 | 0.04 |
ENST00000532032.1
ENST00000506466.1 ENST00000555381.1 ENST00000557311.1 ENST00000554592.1 ENST00000554672.1 ENST00000553491.1 ENST00000556220.1 ENST00000502250.1 ENST00000503767.1 ENST00000393287.5 ENST00000340660.6 ENST00000545170.1 ENST00000429774.2 |
ATXN3
|
ataxin 3 |
| chr9_+_137218362 | 0.04 |
ENST00000481739.1
|
RXRA
|
retinoid X receptor, alpha |
| chr12_+_66217911 | 0.04 |
ENST00000403681.2
|
HMGA2
|
high mobility group AT-hook 2 |
| chr13_+_47127293 | 0.04 |
ENST00000311191.6
|
LRCH1
|
leucine-rich repeats and calponin homology (CH) domain containing 1 |
| chr2_+_202316392 | 0.04 |
ENST00000194530.3
ENST00000392249.2 |
STRADB
|
STE20-related kinase adaptor beta |
| chr5_+_173315283 | 0.04 |
ENST00000265085.5
|
CPEB4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr3_-_142682178 | 0.03 |
ENST00000340634.3
|
PAQR9
|
progestin and adipoQ receptor family member IX |
| chr2_+_169312350 | 0.03 |
ENST00000305747.6
|
CERS6
|
ceramide synthase 6 |
| chrX_-_19988382 | 0.03 |
ENST00000356980.3
ENST00000379687.3 ENST00000379682.4 |
CXorf23
|
chromosome X open reading frame 23 |
| chr1_+_218519577 | 0.03 |
ENST00000366930.4
ENST00000366929.4 |
TGFB2
|
transforming growth factor, beta 2 |
| chr1_+_151584544 | 0.03 |
ENST00000458013.2
ENST00000368843.3 |
SNX27
|
sorting nexin family member 27 |
| chr6_-_24721054 | 0.03 |
ENST00000378119.4
|
C6orf62
|
chromosome 6 open reading frame 62 |
| chr6_+_99282570 | 0.03 |
ENST00000328345.5
|
POU3F2
|
POU class 3 homeobox 2 |
| chr12_+_67663056 | 0.03 |
ENST00000545606.1
|
CAND1
|
cullin-associated and neddylation-dissociated 1 |
| chr5_-_40798263 | 0.03 |
ENST00000296800.4
ENST00000397128.2 |
PRKAA1
|
protein kinase, AMP-activated, alpha 1 catalytic subunit |
| chr1_+_154540246 | 0.03 |
ENST00000368476.3
|
CHRNB2
|
cholinergic receptor, nicotinic, beta 2 (neuronal) |
| chrX_+_154997474 | 0.03 |
ENST00000302805.2
|
SPRY3
|
sprouty homolog 3 (Drosophila) |
| chr1_+_66797687 | 0.03 |
ENST00000371045.5
ENST00000531025.1 ENST00000526197.1 |
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr6_+_41040678 | 0.03 |
ENST00000341376.6
ENST00000353205.5 |
NFYA
|
nuclear transcription factor Y, alpha |
| chr16_-_23521710 | 0.03 |
ENST00000562117.1
ENST00000567468.1 ENST00000562944.1 ENST00000309859.4 |
GGA2
|
golgi-associated, gamma adaptin ear containing, ARF binding protein 2 |
| chr15_+_66679155 | 0.03 |
ENST00000307102.5
|
MAP2K1
|
mitogen-activated protein kinase kinase 1 |
| chr6_+_147525541 | 0.03 |
ENST00000367481.3
ENST00000546097.1 |
STXBP5
|
syntaxin binding protein 5 (tomosyn) |
| chr17_-_76356148 | 0.03 |
ENST00000587578.1
ENST00000330871.2 |
SOCS3
|
suppressor of cytokine signaling 3 |
| chr6_+_107811162 | 0.03 |
ENST00000317357.5
|
SOBP
|
sine oculis binding protein homolog (Drosophila) |
| chr19_+_8483272 | 0.03 |
ENST00000602117.1
|
MARCH2
|
membrane-associated ring finger (C3HC4) 2, E3 ubiquitin protein ligase |
| chr15_-_49338748 | 0.03 |
ENST00000559471.1
|
SECISBP2L
|
SECIS binding protein 2-like |
| chr10_-_74114714 | 0.03 |
ENST00000338820.3
ENST00000394903.2 ENST00000444643.2 |
DNAJB12
|
DnaJ (Hsp40) homolog, subfamily B, member 12 |
| chr2_+_86333301 | 0.03 |
ENST00000254630.7
|
PTCD3
|
pentatricopeptide repeat domain 3 |
| chr1_-_228291136 | 0.03 |
ENST00000272139.4
|
C1orf35
|
chromosome 1 open reading frame 35 |
| chr20_+_11871371 | 0.03 |
ENST00000254977.3
|
BTBD3
|
BTB (POZ) domain containing 3 |
| chr1_+_36273743 | 0.03 |
ENST00000373210.3
|
AGO4
|
argonaute RISC catalytic component 4 |
| chr6_+_163835669 | 0.03 |
ENST00000453779.2
ENST00000275262.7 ENST00000392127.2 ENST00000361752.3 |
QKI
|
QKI, KH domain containing, RNA binding |
| chr5_-_138775177 | 0.03 |
ENST00000302060.5
|
DNAJC18
|
DnaJ (Hsp40) homolog, subfamily C, member 18 |
| chr3_-_195808952 | 0.03 |
ENST00000540528.1
ENST00000392396.3 ENST00000535031.1 ENST00000420415.1 |
TFRC
|
transferrin receptor |
| chr5_+_135468516 | 0.03 |
ENST00000507118.1
ENST00000511116.1 ENST00000545279.1 ENST00000545620.1 |
SMAD5
|
SMAD family member 5 |
| chr11_+_34073195 | 0.03 |
ENST00000341394.4
|
CAPRIN1
|
cell cycle associated protein 1 |
| chr2_+_56411131 | 0.03 |
ENST00000407595.2
|
CCDC85A
|
coiled-coil domain containing 85A |
| chr1_+_68150744 | 0.03 |
ENST00000370986.4
ENST00000370985.3 |
GADD45A
|
growth arrest and DNA-damage-inducible, alpha |
| chr17_+_5031687 | 0.03 |
ENST00000250066.6
ENST00000304328.5 |
USP6
|
ubiquitin specific peptidase 6 (Tre-2 oncogene) |
| chr1_+_93913713 | 0.03 |
ENST00000604705.1
ENST00000370253.2 |
FNBP1L
|
formin binding protein 1-like |
| chr9_+_128024067 | 0.03 |
ENST00000461379.1
ENST00000394084.1 ENST00000394105.2 ENST00000470056.1 ENST00000394104.2 ENST00000265956.4 ENST00000394083.2 ENST00000495955.1 ENST00000467750.1 ENST00000297933.6 |
GAPVD1
|
GTPase activating protein and VPS9 domains 1 |
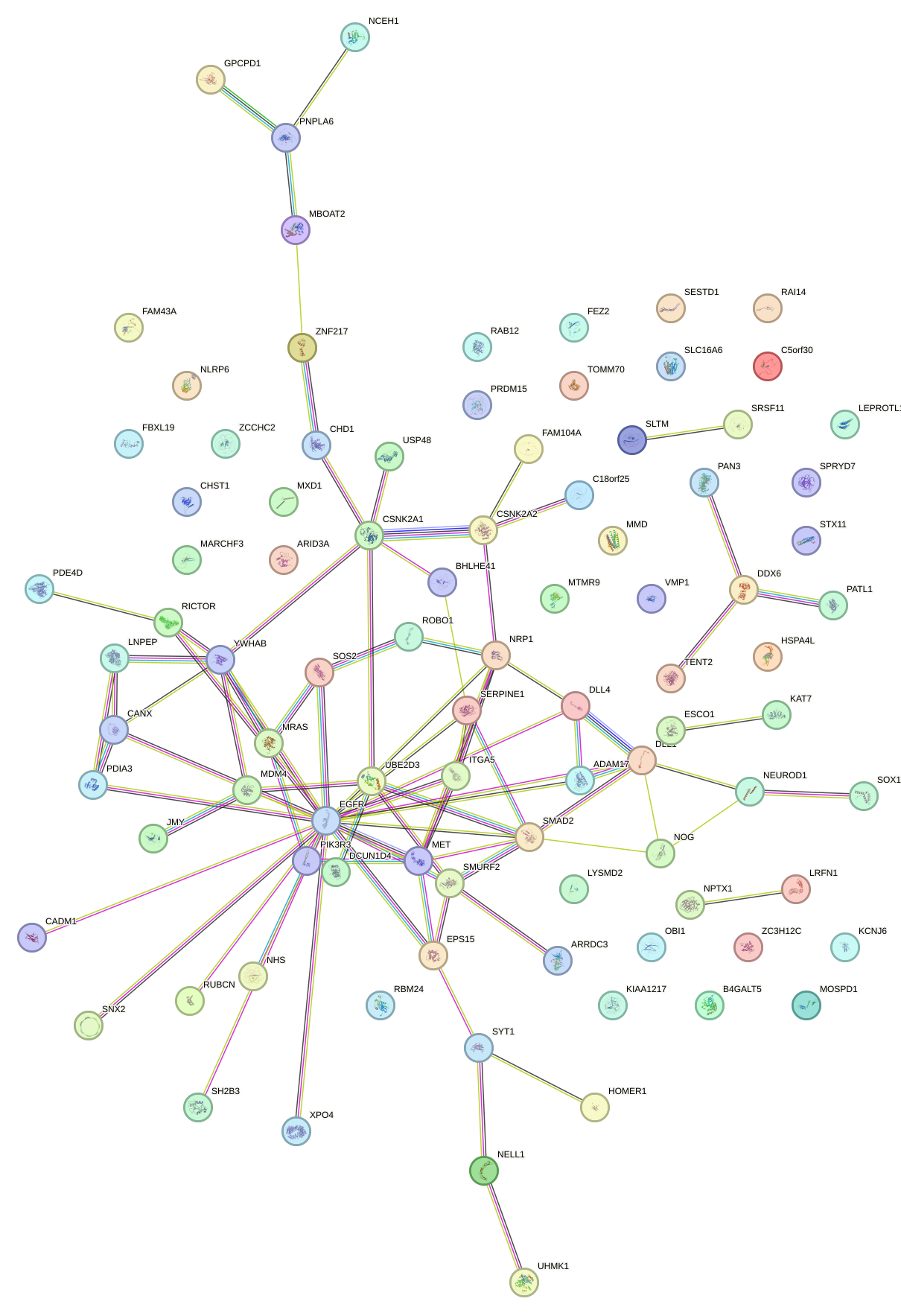
Network of associatons between targets according to the STRING database.
First level regulatory network of CAGUGCA
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0060730 | regulation of intestinal epithelial structure maintenance(GO:0060730) |
| 0.1 | 0.3 | GO:0001300 | chronological cell aging(GO:0001300) |
| 0.1 | 0.2 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.1 | 0.2 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 0.2 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.0 | 0.2 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.0 | 0.2 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.1 | GO:0072720 | response to dithiothreitol(GO:0072720) |
| 0.0 | 0.2 | GO:0075509 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 | 0.1 | GO:0033024 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 0.1 | GO:0048633 | negative regulation of auditory receptor cell differentiation(GO:0045608) negative regulation of mechanoreceptor differentiation(GO:0045632) positive regulation of skeletal muscle tissue growth(GO:0048633) negative regulation of inner ear receptor cell differentiation(GO:2000981) |
| 0.0 | 0.2 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 | 0.1 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.2 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.1 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.1 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 0.0 | 0.1 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.0 | 0.1 | GO:2000973 | midbrain morphogenesis(GO:1904693) regulation of pro-B cell differentiation(GO:2000973) |
| 0.0 | 0.1 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.2 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 0.2 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.1 | GO:0044029 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.0 | 0.1 | GO:0043006 | activation of phospholipase A2 activity by calcium-mediated signaling(GO:0043006) |
| 0.0 | 0.1 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.0 | 0.2 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 0.1 | GO:1990164 | histone H2A phosphorylation(GO:1990164) |
| 0.0 | 0.1 | GO:0072501 | cellular phosphate ion homeostasis(GO:0030643) cellular divalent inorganic anion homeostasis(GO:0072501) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.1 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.0 | 0.3 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.0 | GO:1901254 | modulation by host of viral RNA genome replication(GO:0044830) positive regulation of intracellular transport of viral material(GO:1901254) |
| 0.0 | 0.1 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.0 | 0.2 | GO:0048875 | chemical homeostasis within a tissue(GO:0048875) |
| 0.0 | 0.0 | GO:0045994 | positive regulation of translational initiation by iron(GO:0045994) |
| 0.0 | 0.0 | GO:1904978 | regulation of endosome organization(GO:1904978) |
| 0.0 | 0.1 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 0.0 | GO:0042704 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) uterine wall breakdown(GO:0042704) positive regulation of catagen(GO:0051795) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) |
| 0.0 | 0.1 | GO:0071484 | cellular response to light intensity(GO:0071484) |
| 0.0 | 0.0 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.0 | 0.1 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.0 | 0.0 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.1 | GO:1900060 | negative regulation of sphingolipid biosynthetic process(GO:0090155) negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.0 | 0.2 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 0.1 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.0 | 0.2 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.0 | GO:0032207 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) negative regulation of single strand break repair(GO:1903517) negative regulation of beta-galactosidase activity(GO:1903770) telomere single strand break repair(GO:1903823) negative regulation of telomere single strand break repair(GO:1903824) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.1 | GO:0031379 | RNA-directed RNA polymerase complex(GO:0031379) |
| 0.0 | 0.1 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.0 | 0.2 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.2 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.0 | 0.1 | GO:0097489 | multivesicular body, internal vesicle lumen(GO:0097489) |
| 0.0 | 0.1 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.1 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.0 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.1 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0047389 | glycerophosphocholine phosphodiesterase activity(GO:0047389) |
| 0.0 | 0.2 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.2 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.1 | GO:0043426 | MRF binding(GO:0043426) |
| 0.0 | 0.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.0 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.1 | GO:0030628 | pre-mRNA 3'-splice site binding(GO:0030628) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.0 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.4 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.0 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) mediator complex binding(GO:0036033) |
| 0.0 | 0.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.0 | GO:0047322 | [hydroxymethylglutaryl-CoA reductase (NADPH)] kinase activity(GO:0047322) [acetyl-CoA carboxylase] kinase activity(GO:0050405) |
| 0.0 | 0.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.5 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |