Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
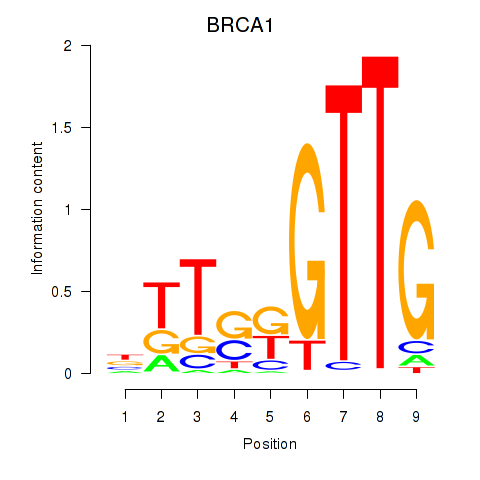
Results for BRCA1
Z-value: 0.21
Transcription factors associated with BRCA1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BRCA1
|
ENSG00000012048.15 | BRCA1 DNA repair associated |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BRCA1 | hg19_v2_chr17_-_41277317_41277348 | 0.55 | 4.5e-01 | Click! |
Activity profile of BRCA1 motif
Sorted Z-values of BRCA1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_7082861 | 0.14 |
ENST00000269299.3
|
ASGR1
|
asialoglycoprotein receptor 1 |
| chr3_-_127455200 | 0.12 |
ENST00000398101.3
|
MGLL
|
monoglyceride lipase |
| chr3_-_176914191 | 0.12 |
ENST00000437738.1
ENST00000424913.1 ENST00000443315.1 |
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1 |
| chr17_-_27230035 | 0.11 |
ENST00000378895.4
ENST00000394901.3 |
DHRS13
|
dehydrogenase/reductase (SDR family) member 13 |
| chr17_+_21730180 | 0.10 |
ENST00000584398.1
|
UBBP4
|
ubiquitin B pseudogene 4 |
| chrX_-_46187069 | 0.09 |
ENST00000446884.1
|
RP1-30G7.2
|
RP1-30G7.2 |
| chr2_-_134326009 | 0.08 |
ENST00000409261.1
ENST00000409213.1 |
NCKAP5
|
NCK-associated protein 5 |
| chr7_+_99717230 | 0.07 |
ENST00000262932.3
|
CNPY4
|
canopy FGF signaling regulator 4 |
| chr3_-_176914238 | 0.07 |
ENST00000430069.1
ENST00000428970.1 |
TBL1XR1
|
transducin (beta)-like 1 X-linked receptor 1 |
| chr19_+_46000479 | 0.06 |
ENST00000456399.2
|
PPM1N
|
protein phosphatase, Mg2+/Mn2+ dependent, 1N (putative) |
| chr11_-_71823266 | 0.06 |
ENST00000538919.1
ENST00000539395.1 ENST00000542531.1 |
ANAPC15
|
anaphase promoting complex subunit 15 |
| chr17_-_56296580 | 0.05 |
ENST00000313863.6
ENST00000546108.1 ENST00000337050.7 ENST00000393119.2 |
MKS1
|
Meckel syndrome, type 1 |
| chr19_+_55888186 | 0.05 |
ENST00000291934.3
|
TMEM190
|
transmembrane protein 190 |
| chr2_+_108994466 | 0.05 |
ENST00000272452.2
|
SULT1C4
|
sulfotransferase family, cytosolic, 1C, member 4 |
| chr19_-_6057282 | 0.05 |
ENST00000592281.1
|
RFX2
|
regulatory factor X, 2 (influences HLA class II expression) |
| chr10_-_69597810 | 0.05 |
ENST00000483798.2
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr10_+_74653330 | 0.05 |
ENST00000334011.5
|
OIT3
|
oncoprotein induced transcript 3 |
| chr8_+_22462532 | 0.04 |
ENST00000389279.3
|
CCAR2
|
cell cycle and apoptosis regulator 2 |
| chr5_+_145583107 | 0.04 |
ENST00000506502.1
|
RBM27
|
RNA binding motif protein 27 |
| chr19_+_45971246 | 0.04 |
ENST00000585836.1
ENST00000417353.2 ENST00000353609.3 ENST00000591858.1 ENST00000443841.2 ENST00000590335.1 |
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B |
| chr19_-_48389651 | 0.04 |
ENST00000222002.3
|
SULT2A1
|
sulfotransferase family, cytosolic, 2A, dehydroepiandrosterone (DHEA)-preferring, member 1 |
| chr4_+_185570767 | 0.04 |
ENST00000314970.6
ENST00000515774.1 ENST00000503752.1 |
PRIMPOL
|
primase and polymerase (DNA-directed) |
| chr3_+_187461442 | 0.04 |
ENST00000450760.1
|
RP11-211G3.2
|
RP11-211G3.2 |
| chr16_-_790887 | 0.04 |
ENST00000540986.1
|
NARFL
|
nuclear prelamin A recognition factor-like |
| chr6_+_42981922 | 0.04 |
ENST00000326974.4
ENST00000244670.8 |
KLHDC3
|
kelch domain containing 3 |
| chr5_+_139027877 | 0.04 |
ENST00000302517.3
|
CXXC5
|
CXXC finger protein 5 |
| chr19_-_37663572 | 0.04 |
ENST00000588354.1
ENST00000292841.5 ENST00000355533.2 ENST00000356958.4 |
ZNF585A
|
zinc finger protein 585A |
| chrX_-_70288234 | 0.04 |
ENST00000276105.3
ENST00000374274.3 |
SNX12
|
sorting nexin 12 |
| chr15_-_88799948 | 0.04 |
ENST00000394480.2
|
NTRK3
|
neurotrophic tyrosine kinase, receptor, type 3 |
| chr10_+_122610687 | 0.04 |
ENST00000263461.6
|
WDR11
|
WD repeat domain 11 |
| chr18_+_55888767 | 0.04 |
ENST00000431212.2
ENST00000586268.1 ENST00000587190.1 |
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr17_+_36584662 | 0.04 |
ENST00000431231.2
ENST00000437668.3 |
ARHGAP23
|
Rho GTPase activating protein 23 |
| chr20_+_9049682 | 0.03 |
ENST00000334005.3
ENST00000378473.3 |
PLCB4
|
phospholipase C, beta 4 |
| chr10_+_104404644 | 0.03 |
ENST00000462202.2
|
TRIM8
|
tripartite motif containing 8 |
| chr10_-_75385711 | 0.03 |
ENST00000433394.1
|
USP54
|
ubiquitin specific peptidase 54 |
| chr10_-_102289611 | 0.03 |
ENST00000299166.4
ENST00000370320.4 ENST00000531258.1 ENST00000370322.1 ENST00000535773.1 |
NDUFB8
SEC31B
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 8, 19kDa SEC31 homolog B (S. cerevisiae) |
| chr3_-_27410847 | 0.03 |
ENST00000429845.2
ENST00000341435.5 ENST00000435750.1 |
NEK10
|
NIMA-related kinase 10 |
| chr12_-_12491608 | 0.03 |
ENST00000545735.1
|
MANSC1
|
MANSC domain containing 1 |
| chr11_+_47291193 | 0.03 |
ENST00000428807.1
ENST00000402799.1 ENST00000406482.1 ENST00000349238.3 ENST00000311027.5 ENST00000407859.3 ENST00000395344.3 ENST00000444117.1 |
MADD
|
MAP-kinase activating death domain |
| chr8_+_97773202 | 0.03 |
ENST00000519484.1
|
CPQ
|
carboxypeptidase Q |
| chr16_-_790982 | 0.03 |
ENST00000301694.5
ENST00000251588.2 |
NARFL
|
nuclear prelamin A recognition factor-like |
| chr19_+_45394477 | 0.03 |
ENST00000252487.5
ENST00000405636.2 ENST00000592434.1 ENST00000426677.2 ENST00000589649.1 |
TOMM40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr6_+_33172407 | 0.03 |
ENST00000374662.3
|
HSD17B8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr12_+_122880045 | 0.03 |
ENST00000539034.1
ENST00000535976.1 |
RP11-450K4.1
|
RP11-450K4.1 |
| chr6_-_170862322 | 0.03 |
ENST00000262193.6
|
PSMB1
|
proteasome (prosome, macropain) subunit, beta type, 1 |
| chr8_+_97773457 | 0.02 |
ENST00000521142.1
|
CPQ
|
carboxypeptidase Q |
| chr2_-_152118276 | 0.02 |
ENST00000409092.1
|
RBM43
|
RNA binding motif protein 43 |
| chr7_+_73868439 | 0.02 |
ENST00000424337.2
|
GTF2IRD1
|
GTF2I repeat domain containing 1 |
| chr11_+_47291645 | 0.02 |
ENST00000395336.3
ENST00000402192.2 |
MADD
|
MAP-kinase activating death domain |
| chr2_+_203879568 | 0.02 |
ENST00000449802.1
|
NBEAL1
|
neurobeachin-like 1 |
| chr2_-_152118352 | 0.02 |
ENST00000331426.5
|
RBM43
|
RNA binding motif protein 43 |
| chr5_+_139493665 | 0.02 |
ENST00000331327.3
|
PURA
|
purine-rich element binding protein A |
| chr2_+_196521458 | 0.02 |
ENST00000409086.3
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr1_+_10271674 | 0.02 |
ENST00000377086.1
|
KIF1B
|
kinesin family member 1B |
| chrX_+_150565038 | 0.02 |
ENST00000370361.1
|
VMA21
|
VMA21 vacuolar H+-ATPase homolog (S. cerevisiae) |
| chr6_+_123100620 | 0.02 |
ENST00000368444.3
|
FABP7
|
fatty acid binding protein 7, brain |
| chr9_+_35792151 | 0.01 |
ENST00000342694.2
|
NPR2
|
natriuretic peptide receptor B/guanylate cyclase B (atrionatriuretic peptide receptor B) |
| chr4_+_619386 | 0.01 |
ENST00000496514.1
|
PDE6B
|
phosphodiesterase 6B, cGMP-specific, rod, beta |
| chr4_+_619347 | 0.01 |
ENST00000255622.6
|
PDE6B
|
phosphodiesterase 6B, cGMP-specific, rod, beta |
| chr12_-_95510743 | 0.01 |
ENST00000551521.1
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6 |
| chr12_-_117319236 | 0.01 |
ENST00000257572.5
|
HRK
|
harakiri, BCL2 interacting protein (contains only BH3 domain) |
| chr15_-_72612470 | 0.01 |
ENST00000287202.5
|
CELF6
|
CUGBP, Elav-like family member 6 |
| chr11_-_27722021 | 0.01 |
ENST00000356660.4
ENST00000418212.1 ENST00000533246.1 |
BDNF
|
brain-derived neurotrophic factor |
| chr8_-_116681123 | 0.01 |
ENST00000519674.1
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr6_-_42981651 | 0.01 |
ENST00000244711.3
|
MEA1
|
male-enhanced antigen 1 |
| chr2_+_223725652 | 0.01 |
ENST00000357430.3
ENST00000392066.3 |
ACSL3
|
acyl-CoA synthetase long-chain family member 3 |
| chr12_+_510795 | 0.01 |
ENST00000412006.2
|
CCDC77
|
coiled-coil domain containing 77 |
| chr1_-_227505289 | 0.01 |
ENST00000366765.3
|
CDC42BPA
|
CDC42 binding protein kinase alpha (DMPK-like) |
| chr13_+_27825446 | 0.01 |
ENST00000311549.6
|
RPL21
|
ribosomal protein L21 |
| chr12_+_510742 | 0.01 |
ENST00000239830.4
|
CCDC77
|
coiled-coil domain containing 77 |
| chr7_-_99716952 | 0.01 |
ENST00000523306.1
ENST00000344095.4 ENST00000417349.1 ENST00000493322.1 ENST00000520135.1 ENST00000418432.2 ENST00000460673.2 ENST00000452041.1 ENST00000452438.2 ENST00000451699.1 ENST00000453269.2 |
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr11_+_117947782 | 0.01 |
ENST00000522307.1
ENST00000523251.1 ENST00000437212.3 ENST00000522824.1 ENST00000522151.1 |
TMPRSS4
|
transmembrane protease, serine 4 |
| chr7_-_78400598 | 0.01 |
ENST00000536571.1
|
MAGI2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr12_-_122879969 | 0.01 |
ENST00000540304.1
|
CLIP1
|
CAP-GLY domain containing linker protein 1 |
| chr8_-_81083341 | 0.01 |
ENST00000519303.2
|
TPD52
|
tumor protein D52 |
| chr1_+_161736072 | 0.00 |
ENST00000367942.3
|
ATF6
|
activating transcription factor 6 |
| chr6_-_137314371 | 0.00 |
ENST00000432330.1
ENST00000418699.1 |
RP11-55K22.5
|
RP11-55K22.5 |
| chr10_-_61900762 | 0.00 |
ENST00000355288.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr17_-_60142609 | 0.00 |
ENST00000397786.2
|
MED13
|
mediator complex subunit 13 |
| chr5_+_145583156 | 0.00 |
ENST00000265271.5
|
RBM27
|
RNA binding motif protein 27 |
| chr2_+_61404624 | 0.00 |
ENST00000394457.3
|
AHSA2
|
AHA1, activator of heat shock 90kDa protein ATPase homolog 2 (yeast) |
| chr4_-_140005443 | 0.00 |
ENST00000510408.1
ENST00000420916.2 ENST00000358635.3 |
ELF2
|
E74-like factor 2 (ets domain transcription factor) |
| chr7_+_44646177 | 0.00 |
ENST00000443864.2
ENST00000447398.1 ENST00000449767.1 ENST00000419661.1 |
OGDH
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) |
| chr19_+_46850320 | 0.00 |
ENST00000391919.1
|
PPP5C
|
protein phosphatase 5, catalytic subunit |
| chr10_+_122610853 | 0.00 |
ENST00000604585.1
|
WDR11
|
WD repeat domain 11 |
| chr12_-_49449107 | 0.00 |
ENST00000301067.7
|
KMT2D
|
lysine (K)-specific methyltransferase 2D |
| chr9_-_86593238 | 0.00 |
ENST00000351839.3
|
HNRNPK
|
heterogeneous nuclear ribonucleoprotein K |
| chr17_+_36908984 | 0.00 |
ENST00000225426.4
ENST00000579088.1 |
PSMB3
|
proteasome (prosome, macropain) subunit, beta type, 3 |
| chr12_-_96336369 | 0.00 |
ENST00000546947.1
ENST00000546386.1 |
CCDC38
|
coiled-coil domain containing 38 |
| chr1_-_160232197 | 0.00 |
ENST00000419626.1
ENST00000610139.1 ENST00000475733.1 ENST00000407642.2 ENST00000368073.3 ENST00000326837.2 |
DCAF8
|
DDB1 and CUL4 associated factor 8 |
| chr11_-_67141640 | 0.00 |
ENST00000533438.1
|
CLCF1
|
cardiotrophin-like cytokine factor 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of BRCA1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 0.2 | GO:0060613 | fat pad development(GO:0060613) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.0 | 0.1 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.0 | GO:0003896 | DNA primase activity(GO:0003896) |