Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
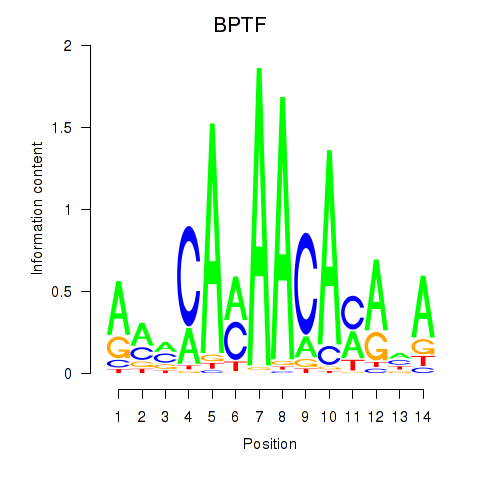
Results for BPTF
Z-value: 1.16
Transcription factors associated with BPTF
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BPTF
|
ENSG00000171634.12 | bromodomain PHD finger transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BPTF | hg19_v2_chr17_+_65821636_65821641 | 0.76 | 2.4e-01 | Click! |
Activity profile of BPTF motif
Sorted Z-values of BPTF motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_-_37976082 | 0.78 |
ENST00000215886.4
|
LGALS2
|
lectin, galactoside-binding, soluble, 2 |
| chr14_-_55658323 | 0.68 |
ENST00000554067.1
ENST00000247191.2 |
DLGAP5
|
discs, large (Drosophila) homolog-associated protein 5 |
| chr4_-_74847800 | 0.67 |
ENST00000296029.3
|
PF4
|
platelet factor 4 |
| chr5_+_82767583 | 0.66 |
ENST00000512590.2
ENST00000513960.1 ENST00000513984.1 ENST00000502527.2 |
VCAN
|
versican |
| chr15_-_31521567 | 0.55 |
ENST00000560812.1
ENST00000559853.1 ENST00000558109.1 |
RP11-16E12.2
|
RP11-16E12.2 |
| chr14_-_45722360 | 0.51 |
ENST00000451174.1
|
MIS18BP1
|
MIS18 binding protein 1 |
| chr17_+_26638667 | 0.49 |
ENST00000600595.1
|
AC061975.10
|
Uncharacterized protein |
| chr5_+_162932554 | 0.47 |
ENST00000321757.6
ENST00000421814.2 ENST00000518095.1 |
MAT2B
|
methionine adenosyltransferase II, beta |
| chr18_+_29171689 | 0.46 |
ENST00000237014.3
|
TTR
|
transthyretin |
| chr15_+_55611128 | 0.45 |
ENST00000164305.5
ENST00000539642.1 |
PIGB
|
phosphatidylinositol glycan anchor biosynthesis, class B |
| chr12_+_69753448 | 0.43 |
ENST00000247843.2
ENST00000548020.1 ENST00000549685.1 ENST00000552955.1 |
YEATS4
|
YEATS domain containing 4 |
| chr11_-_69867159 | 0.42 |
ENST00000528507.1
|
RP11-626H12.2
|
RP11-626H12.2 |
| chr6_+_26104104 | 0.42 |
ENST00000377803.2
|
HIST1H4C
|
histone cluster 1, H4c |
| chr9_+_67977438 | 0.42 |
ENST00000456982.1
|
RP11-195B21.3
|
Protein LOC644249 |
| chr5_+_82767487 | 0.41 |
ENST00000343200.5
ENST00000342785.4 |
VCAN
|
versican |
| chr19_-_28283939 | 0.41 |
ENST00000591338.1
|
LINC00662
|
long intergenic non-protein coding RNA 662 |
| chr15_+_55611401 | 0.41 |
ENST00000566999.1
|
PIGB
|
phosphatidylinositol glycan anchor biosynthesis, class B |
| chr1_-_220263096 | 0.40 |
ENST00000463953.1
ENST00000354807.3 ENST00000414869.2 ENST00000498237.2 ENST00000498791.2 ENST00000544404.1 ENST00000480959.2 ENST00000322067.7 |
BPNT1
|
3'(2'), 5'-bisphosphate nucleotidase 1 |
| chr15_-_55657428 | 0.39 |
ENST00000568543.1
|
CCPG1
|
cell cycle progression 1 |
| chr12_-_56223363 | 0.39 |
ENST00000546957.1
|
DNAJC14
|
DnaJ (Hsp40) homolog, subfamily C, member 14 |
| chr3_+_93698974 | 0.39 |
ENST00000535334.1
ENST00000478400.1 ENST00000303097.7 ENST00000394222.3 ENST00000471138.1 ENST00000539730.1 |
ARL13B
|
ADP-ribosylation factor-like 13B |
| chr17_-_4938712 | 0.38 |
ENST00000254853.5
ENST00000424747.1 |
SLC52A1
|
solute carrier family 52 (riboflavin transporter), member 1 |
| chr8_-_83589388 | 0.37 |
ENST00000522776.1
|
RP11-653B10.1
|
RP11-653B10.1 |
| chr11_+_107643129 | 0.36 |
ENST00000447610.1
|
AP001024.2
|
Uncharacterized protein |
| chr19_-_36297348 | 0.35 |
ENST00000589835.1
|
PRODH2
|
proline dehydrogenase (oxidase) 2 |
| chr7_-_143599207 | 0.34 |
ENST00000355951.2
ENST00000479870.1 ENST00000478172.1 |
FAM115A
|
family with sequence similarity 115, member A |
| chr19_-_42192189 | 0.34 |
ENST00000401731.1
ENST00000338196.4 ENST00000006724.3 |
CEACAM7
|
carcinoembryonic antigen-related cell adhesion molecule 7 |
| chr22_-_30901637 | 0.33 |
ENST00000381982.3
ENST00000255858.7 ENST00000540456.1 ENST00000392772.2 |
SEC14L4
|
SEC14-like 4 (S. cerevisiae) |
| chr14_-_80678512 | 0.33 |
ENST00000553968.1
|
DIO2
|
deiodinase, iodothyronine, type II |
| chr9_+_42671887 | 0.33 |
ENST00000456520.1
ENST00000377391.3 |
CBWD7
|
COBW domain containing 7 |
| chr21_-_35340759 | 0.33 |
ENST00000607953.1
|
AP000569.9
|
AP000569.9 |
| chr17_-_62097927 | 0.32 |
ENST00000578313.1
ENST00000584084.1 ENST00000579788.1 ENST00000579687.1 ENST00000578379.1 ENST00000578892.1 ENST00000412356.1 ENST00000418105.1 |
ICAM2
|
intercellular adhesion molecule 2 |
| chr3_+_149192475 | 0.32 |
ENST00000465758.1
|
TM4SF4
|
transmembrane 4 L six family member 4 |
| chr6_-_15548591 | 0.32 |
ENST00000509674.1
|
DTNBP1
|
dystrobrevin binding protein 1 |
| chr19_-_45457264 | 0.32 |
ENST00000591646.1
|
CTB-129P6.11
|
Uncharacterized protein |
| chrX_-_77041685 | 0.32 |
ENST00000373344.5
ENST00000395603.3 |
ATRX
|
alpha thalassemia/mental retardation syndrome X-linked |
| chrY_+_24636544 | 0.31 |
ENST00000303728.1
ENST00000338793.2 |
PRY
|
PTPN13-like, Y-linked |
| chr16_+_53483983 | 0.31 |
ENST00000544545.1
|
RBL2
|
retinoblastoma-like 2 (p130) |
| chr8_-_95274536 | 0.31 |
ENST00000297596.2
ENST00000396194.2 |
GEM
|
GTP binding protein overexpressed in skeletal muscle |
| chr11_+_119056178 | 0.31 |
ENST00000525131.1
ENST00000531114.1 ENST00000355547.5 ENST00000322712.4 |
PDZD3
|
PDZ domain containing 3 |
| chr1_+_59486129 | 0.31 |
ENST00000438195.1
ENST00000424308.1 |
RP4-794H19.4
|
RP4-794H19.4 |
| chr10_+_63422695 | 0.31 |
ENST00000330194.2
ENST00000389639.3 |
C10orf107
|
chromosome 10 open reading frame 107 |
| chr12_-_57146095 | 0.31 |
ENST00000550770.1
ENST00000338193.6 |
PRIM1
|
primase, DNA, polypeptide 1 (49kDa) |
| chr6_-_166309725 | 0.30 |
ENST00000545867.1
|
SDIM1
|
stress responsive DNAJB4 interacting membrane protein 1 |
| chr1_+_93297622 | 0.30 |
ENST00000315741.5
|
RPL5
|
ribosomal protein L5 |
| chrY_-_24242154 | 0.30 |
ENST00000303804.1
ENST00000341740.2 |
PRY2
|
PTPN13-like, Y-linked 2 |
| chr4_+_89300158 | 0.29 |
ENST00000502870.1
|
HERC6
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 6 |
| chr9_+_2158443 | 0.29 |
ENST00000302401.3
ENST00000324954.5 ENST00000423555.1 ENST00000382186.1 |
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr17_-_74351010 | 0.29 |
ENST00000435555.2
|
PRPSAP1
|
phosphoribosyl pyrophosphate synthetase-associated protein 1 |
| chr8_-_27941380 | 0.29 |
ENST00000413272.2
ENST00000341513.6 |
NUGGC
|
nuclear GTPase, germinal center associated |
| chr16_+_53920795 | 0.29 |
ENST00000431610.2
ENST00000460382.1 |
FTO
|
fat mass and obesity associated |
| chr22_-_42336209 | 0.29 |
ENST00000472374.2
|
CENPM
|
centromere protein M |
| chr5_+_71475449 | 0.29 |
ENST00000504492.1
|
MAP1B
|
microtubule-associated protein 1B |
| chr1_+_64014588 | 0.29 |
ENST00000371086.2
ENST00000340052.3 |
DLEU2L
|
deleted in lymphocytic leukemia 2-like |
| chr6_+_76599809 | 0.29 |
ENST00000430435.1
|
MYO6
|
myosin VI |
| chr6_-_107230334 | 0.29 |
ENST00000607090.1
|
RP1-60O19.1
|
RP1-60O19.1 |
| chr19_-_58204128 | 0.29 |
ENST00000597520.1
|
AC004017.1
|
Uncharacterized protein |
| chr4_+_96012614 | 0.28 |
ENST00000264568.4
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr8_+_97597148 | 0.28 |
ENST00000521590.1
|
SDC2
|
syndecan 2 |
| chr13_+_44596471 | 0.28 |
ENST00000592085.1
ENST00000591799.1 |
LINC00284
|
long intergenic non-protein coding RNA 284 |
| chr1_-_170043709 | 0.28 |
ENST00000367767.1
ENST00000361580.2 ENST00000538366.1 |
KIFAP3
|
kinesin-associated protein 3 |
| chr4_-_88312301 | 0.28 |
ENST00000507286.1
|
HSD17B11
|
hydroxysteroid (17-beta) dehydrogenase 11 |
| chr16_+_21623958 | 0.28 |
ENST00000568826.1
|
METTL9
|
methyltransferase like 9 |
| chr5_-_34919094 | 0.28 |
ENST00000341754.4
|
RAD1
|
RAD1 homolog (S. pombe) |
| chr17_+_21729899 | 0.27 |
ENST00000583708.1
|
UBBP4
|
ubiquitin B pseudogene 4 |
| chr19_-_18197714 | 0.27 |
ENST00000322153.7
|
IL12RB1
|
interleukin 12 receptor, beta 1 |
| chr16_-_28621353 | 0.27 |
ENST00000567512.1
|
SULT1A1
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr17_-_64225508 | 0.27 |
ENST00000205948.6
|
APOH
|
apolipoprotein H (beta-2-glycoprotein I) |
| chr9_+_92219919 | 0.27 |
ENST00000252506.6
ENST00000375769.1 |
GADD45G
|
growth arrest and DNA-damage-inducible, gamma |
| chr5_+_138611798 | 0.26 |
ENST00000502394.1
|
MATR3
|
matrin 3 |
| chr3_-_178969403 | 0.26 |
ENST00000314235.5
ENST00000392685.2 |
KCNMB3
|
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr15_+_32933866 | 0.26 |
ENST00000300175.4
ENST00000413748.2 ENST00000494364.1 ENST00000497208.1 |
SCG5
|
secretogranin V (7B2 protein) |
| chr1_-_197036364 | 0.26 |
ENST00000367412.1
|
F13B
|
coagulation factor XIII, B polypeptide |
| chr4_+_157997273 | 0.26 |
ENST00000541722.1
ENST00000512619.1 |
GLRB
|
glycine receptor, beta |
| chr12_-_121410095 | 0.26 |
ENST00000539163.1
|
AC079602.1
|
AC079602.1 |
| chr12_-_123728548 | 0.26 |
ENST00000545406.1
|
MPHOSPH9
|
M-phase phosphoprotein 9 |
| chr3_+_127323739 | 0.25 |
ENST00000491422.1
|
MCM2
|
minichromosome maintenance complex component 2 |
| chr17_-_7080227 | 0.25 |
ENST00000574330.1
|
ASGR1
|
asialoglycoprotein receptor 1 |
| chr14_-_55658252 | 0.25 |
ENST00000395425.2
|
DLGAP5
|
discs, large (Drosophila) homolog-associated protein 5 |
| chr16_-_28550348 | 0.25 |
ENST00000324873.6
|
NUPR1
|
nuclear protein, transcriptional regulator, 1 |
| chr17_-_10633291 | 0.25 |
ENST00000578345.1
ENST00000455996.2 |
TMEM220
|
transmembrane protein 220 |
| chr10_-_101841588 | 0.25 |
ENST00000370418.3
|
CPN1
|
carboxypeptidase N, polypeptide 1 |
| chr1_-_193075180 | 0.25 |
ENST00000367440.3
|
GLRX2
|
glutaredoxin 2 |
| chr12_-_89918982 | 0.25 |
ENST00000549504.1
|
POC1B
|
POC1 centriolar protein B |
| chr9_+_2158485 | 0.25 |
ENST00000417599.1
ENST00000382185.1 ENST00000382183.1 |
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr5_+_162864575 | 0.25 |
ENST00000512163.1
ENST00000393929.1 ENST00000340828.2 ENST00000511683.2 ENST00000510097.1 ENST00000511490.2 ENST00000510664.1 |
CCNG1
|
cyclin G1 |
| chr3_+_158362299 | 0.25 |
ENST00000478576.1
ENST00000264263.5 ENST00000464732.1 |
GFM1
|
G elongation factor, mitochondrial 1 |
| chr1_+_28261492 | 0.25 |
ENST00000373894.3
|
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chrX_+_106163626 | 0.25 |
ENST00000336803.1
|
CLDN2
|
claudin 2 |
| chr4_+_165675197 | 0.25 |
ENST00000515485.1
|
RP11-294O2.2
|
RP11-294O2.2 |
| chr3_-_139396801 | 0.24 |
ENST00000413939.2
ENST00000339837.5 ENST00000512391.1 |
NMNAT3
|
nicotinamide nucleotide adenylyltransferase 3 |
| chr2_-_36779411 | 0.24 |
ENST00000406220.1
|
AC007401.2
|
Uncharacterized protein |
| chr15_+_66585555 | 0.24 |
ENST00000319194.5
ENST00000525134.2 ENST00000441424.2 |
DIS3L
|
DIS3 mitotic control homolog (S. cerevisiae)-like |
| chr3_-_127455200 | 0.24 |
ENST00000398101.3
|
MGLL
|
monoglyceride lipase |
| chr6_+_80341000 | 0.24 |
ENST00000369838.4
|
SH3BGRL2
|
SH3 domain binding glutamic acid-rich protein like 2 |
| chr3_-_112360116 | 0.24 |
ENST00000206423.3
ENST00000439685.2 |
CCDC80
|
coiled-coil domain containing 80 |
| chr3_+_186330712 | 0.24 |
ENST00000411641.2
ENST00000273784.5 |
AHSG
|
alpha-2-HS-glycoprotein |
| chr4_+_72204755 | 0.23 |
ENST00000512686.1
ENST00000340595.3 |
SLC4A4
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
| chr4_-_155511887 | 0.23 |
ENST00000302053.3
ENST00000403106.3 |
FGA
|
fibrinogen alpha chain |
| chr6_+_26087509 | 0.23 |
ENST00000397022.3
ENST00000353147.5 ENST00000352392.4 ENST00000349999.4 ENST00000317896.7 ENST00000357618.5 ENST00000470149.1 ENST00000336625.8 ENST00000461397.1 ENST00000488199.1 |
HFE
|
hemochromatosis |
| chr14_-_64108125 | 0.23 |
ENST00000267522.3
|
WDR89
|
WD repeat domain 89 |
| chr9_-_123812542 | 0.23 |
ENST00000223642.1
|
C5
|
complement component 5 |
| chr9_+_5510492 | 0.23 |
ENST00000397745.2
|
PDCD1LG2
|
programmed cell death 1 ligand 2 |
| chr19_-_10613862 | 0.23 |
ENST00000592055.1
|
KEAP1
|
kelch-like ECH-associated protein 1 |
| chr3_+_187461442 | 0.23 |
ENST00000450760.1
|
RP11-211G3.2
|
RP11-211G3.2 |
| chrX_+_66764375 | 0.23 |
ENST00000374690.3
|
AR
|
androgen receptor |
| chr9_-_47314222 | 0.22 |
ENST00000420228.1
ENST00000438517.1 ENST00000414020.1 |
AL953854.2
|
AL953854.2 |
| chr20_-_17539456 | 0.22 |
ENST00000544874.1
ENST00000377868.2 |
BFSP1
|
beaded filament structural protein 1, filensin |
| chr8_+_27631903 | 0.22 |
ENST00000305188.8
|
ESCO2
|
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr16_-_28550320 | 0.22 |
ENST00000395641.2
|
NUPR1
|
nuclear protein, transcriptional regulator, 1 |
| chr2_+_33359687 | 0.22 |
ENST00000402934.1
ENST00000404525.1 ENST00000407925.1 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr2_+_102686820 | 0.22 |
ENST00000409929.1
ENST00000424272.1 |
IL1R1
|
interleukin 1 receptor, type I |
| chr3_+_118892362 | 0.22 |
ENST00000497685.1
ENST00000264234.3 |
UPK1B
|
uroplakin 1B |
| chr1_-_150207017 | 0.22 |
ENST00000369119.3
|
ANP32E
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr3_-_56835967 | 0.22 |
ENST00000495373.1
ENST00000296315.3 |
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3 |
| chr1_+_10292308 | 0.22 |
ENST00000377081.1
|
KIF1B
|
kinesin family member 1B |
| chr7_-_35013217 | 0.22 |
ENST00000446375.1
|
DPY19L1
|
dpy-19-like 1 (C. elegans) |
| chr9_+_5510558 | 0.22 |
ENST00000397747.3
|
PDCD1LG2
|
programmed cell death 1 ligand 2 |
| chr2_-_68290106 | 0.22 |
ENST00000407324.1
ENST00000355848.3 ENST00000409302.1 ENST00000410067.3 |
C1D
|
C1D nuclear receptor corepressor |
| chr2_-_190445499 | 0.22 |
ENST00000261024.2
|
SLC40A1
|
solute carrier family 40 (iron-regulated transporter), member 1 |
| chr3_+_42632298 | 0.22 |
ENST00000011691.4
|
SS18L2
|
synovial sarcoma translocation gene on chromosome 18-like 2 |
| chr19_-_47735918 | 0.22 |
ENST00000449228.1
ENST00000300880.7 ENST00000341983.4 |
BBC3
|
BCL2 binding component 3 |
| chr5_-_159739483 | 0.22 |
ENST00000519673.1
ENST00000541762.1 |
CCNJL
|
cyclin J-like |
| chr7_-_73256838 | 0.21 |
ENST00000297873.4
|
WBSCR27
|
Williams Beuren syndrome chromosome region 27 |
| chr9_-_6015607 | 0.21 |
ENST00000259569.5
|
RANBP6
|
RAN binding protein 6 |
| chr14_+_56127960 | 0.21 |
ENST00000553624.1
|
KTN1
|
kinectin 1 (kinesin receptor) |
| chr7_+_23210760 | 0.21 |
ENST00000366347.4
|
AC005082.1
|
Uncharacterized protein |
| chr8_-_27457494 | 0.21 |
ENST00000521770.1
|
CLU
|
clusterin |
| chr21_+_17566643 | 0.21 |
ENST00000419952.1
ENST00000445461.2 |
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr17_+_42219267 | 0.21 |
ENST00000319977.4
ENST00000585683.1 |
C17orf53
|
chromosome 17 open reading frame 53 |
| chr6_+_37897735 | 0.21 |
ENST00000373389.5
|
ZFAND3
|
zinc finger, AN1-type domain 3 |
| chr19_-_36304201 | 0.21 |
ENST00000301175.3
|
PRODH2
|
proline dehydrogenase (oxidase) 2 |
| chr1_+_241695424 | 0.20 |
ENST00000366558.3
ENST00000366559.4 |
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr19_-_4043154 | 0.20 |
ENST00000535853.1
|
AC016586.1
|
AC016586.1 |
| chr12_+_56114151 | 0.20 |
ENST00000547072.1
ENST00000552930.1 ENST00000257895.5 |
RDH5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr2_-_169769787 | 0.20 |
ENST00000451987.1
|
SPC25
|
SPC25, NDC80 kinetochore complex component |
| chr6_+_125524785 | 0.20 |
ENST00000392482.2
|
TPD52L1
|
tumor protein D52-like 1 |
| chr2_+_33359646 | 0.20 |
ENST00000390003.4
ENST00000418533.2 |
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr15_+_30918879 | 0.20 |
ENST00000428041.2
|
ARHGAP11B
|
Rho GTPase activating protein 11B |
| chr17_+_21730180 | 0.20 |
ENST00000584398.1
|
UBBP4
|
ubiquitin B pseudogene 4 |
| chr1_+_241695670 | 0.20 |
ENST00000366557.4
|
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr2_+_38177575 | 0.20 |
ENST00000407257.1
ENST00000417700.2 ENST00000234195.3 ENST00000442857.1 |
RMDN2
|
regulator of microtubule dynamics 2 |
| chr2_-_3523507 | 0.20 |
ENST00000327435.6
|
ADI1
|
acireductone dioxygenase 1 |
| chr7_+_151771377 | 0.20 |
ENST00000434507.1
|
GALNT11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 (GalNAc-T11) |
| chr17_+_42219383 | 0.20 |
ENST00000245382.6
|
C17orf53
|
chromosome 17 open reading frame 53 |
| chr1_+_28261621 | 0.20 |
ENST00000549094.1
|
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr9_-_14313893 | 0.20 |
ENST00000380921.3
ENST00000380959.3 |
NFIB
|
nuclear factor I/B |
| chr8_+_87111059 | 0.20 |
ENST00000285393.3
|
ATP6V0D2
|
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2 |
| chr15_-_76304731 | 0.20 |
ENST00000394907.3
|
NRG4
|
neuregulin 4 |
| chr3_-_139396853 | 0.19 |
ENST00000406164.1
ENST00000406824.1 |
NMNAT3
|
nicotinamide nucleotide adenylyltransferase 3 |
| chr9_+_40028620 | 0.19 |
ENST00000426179.1
|
AL353791.1
|
AL353791.1 |
| chr15_+_35270552 | 0.19 |
ENST00000391457.2
|
AC114546.1
|
HCG37415; PRO1914; Uncharacterized protein |
| chr4_+_174818390 | 0.19 |
ENST00000509968.1
ENST00000512263.1 |
RP11-161D15.1
|
RP11-161D15.1 |
| chr7_-_16840820 | 0.19 |
ENST00000450569.1
|
AGR2
|
anterior gradient 2 |
| chr3_+_101292939 | 0.19 |
ENST00000265260.3
ENST00000469941.1 ENST00000296024.5 |
PCNP
|
PEST proteolytic signal containing nuclear protein |
| chr5_-_54603368 | 0.19 |
ENST00000508346.1
ENST00000251636.5 |
DHX29
|
DEAH (Asp-Glu-Ala-His) box polypeptide 29 |
| chr17_+_21729593 | 0.19 |
ENST00000581769.1
ENST00000584755.1 |
UBBP4
|
ubiquitin B pseudogene 4 |
| chr1_+_28261533 | 0.19 |
ENST00000411604.1
ENST00000373888.4 |
SMPDL3B
|
sphingomyelin phosphodiesterase, acid-like 3B |
| chr16_+_10479906 | 0.19 |
ENST00000562527.1
ENST00000396560.2 ENST00000396559.1 ENST00000562102.1 ENST00000543967.1 ENST00000569939.1 ENST00000569900.1 |
ATF7IP2
|
activating transcription factor 7 interacting protein 2 |
| chr2_-_152118276 | 0.19 |
ENST00000409092.1
|
RBM43
|
RNA binding motif protein 43 |
| chr4_-_153303658 | 0.19 |
ENST00000296555.5
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr12_-_110906027 | 0.19 |
ENST00000537466.2
ENST00000550974.1 ENST00000228827.3 |
GPN3
|
GPN-loop GTPase 3 |
| chr1_+_95699704 | 0.19 |
ENST00000370202.4
|
RWDD3
|
RWD domain containing 3 |
| chr12_-_51418549 | 0.18 |
ENST00000548150.1
|
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr12_+_58166726 | 0.18 |
ENST00000546504.1
|
RP11-571M6.15
|
Uncharacterized protein |
| chr4_+_141264597 | 0.18 |
ENST00000338517.4
ENST00000394203.3 ENST00000506322.1 |
SCOC
|
short coiled-coil protein |
| chr15_+_96876340 | 0.18 |
ENST00000453270.2
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr1_-_65533390 | 0.18 |
ENST00000448344.1
|
RP4-535B20.1
|
RP4-535B20.1 |
| chr17_-_74547256 | 0.18 |
ENST00000589145.1
|
CYGB
|
cytoglobin |
| chr16_-_79804394 | 0.18 |
ENST00000567993.1
|
RP11-345M22.2
|
RP11-345M22.2 |
| chr1_-_149459549 | 0.18 |
ENST00000369175.3
|
FAM72C
|
family with sequence similarity 72, member C |
| chr1_-_165667545 | 0.18 |
ENST00000538148.1
|
ALDH9A1
|
aldehyde dehydrogenase 9 family, member A1 |
| chr6_+_150690133 | 0.18 |
ENST00000392255.3
ENST00000500320.3 |
IYD
|
iodotyrosine deiodinase |
| chr3_+_44840679 | 0.18 |
ENST00000425755.1
|
KIF15
|
kinesin family member 15 |
| chr7_-_16844611 | 0.18 |
ENST00000401412.1
ENST00000419304.2 |
AGR2
|
anterior gradient 2 |
| chr4_+_157997209 | 0.18 |
ENST00000264428.4
|
GLRB
|
glycine receptor, beta |
| chr9_-_36276966 | 0.18 |
ENST00000543356.2
ENST00000396594.3 |
GNE
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
| chr9_+_2157655 | 0.18 |
ENST00000452193.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr1_+_214776516 | 0.18 |
ENST00000366955.3
|
CENPF
|
centromere protein F, 350/400kDa |
| chr6_-_138428613 | 0.18 |
ENST00000421351.3
|
PERP
|
PERP, TP53 apoptosis effector |
| chr22_+_29168652 | 0.18 |
ENST00000249064.4
ENST00000444523.1 ENST00000448492.2 ENST00000421503.2 |
CCDC117
|
coiled-coil domain containing 117 |
| chr19_-_6379069 | 0.18 |
ENST00000597721.1
|
PSPN
|
persephin |
| chr1_+_45212051 | 0.18 |
ENST00000372222.3
|
KIF2C
|
kinesin family member 2C |
| chr10_-_118885954 | 0.18 |
ENST00000392901.4
|
KIAA1598
|
KIAA1598 |
| chr11_-_112034803 | 0.18 |
ENST00000528832.1
|
IL18
|
interleukin 18 (interferon-gamma-inducing factor) |
| chr19_-_33360647 | 0.18 |
ENST00000590341.1
ENST00000587772.1 ENST00000023064.4 |
SLC7A9
|
solute carrier family 7 (amino acid transporter light chain, bo,+ system), member 9 |
| chr17_-_46692457 | 0.18 |
ENST00000468443.1
|
HOXB8
|
homeobox B8 |
| chr12_-_39734783 | 0.18 |
ENST00000552961.1
|
KIF21A
|
kinesin family member 21A |
| chr5_-_42811986 | 0.17 |
ENST00000511224.1
ENST00000507920.1 ENST00000510965.1 |
SEPP1
|
selenoprotein P, plasma, 1 |
| chr5_-_159846066 | 0.17 |
ENST00000519349.1
ENST00000520664.1 |
SLU7
|
SLU7 splicing factor homolog (S. cerevisiae) |
| chr1_+_154377669 | 0.17 |
ENST00000368485.3
ENST00000344086.4 |
IL6R
|
interleukin 6 receptor |
| chr19_+_29704142 | 0.17 |
ENST00000587859.1
ENST00000590607.1 |
CTB-32O4.2
|
CTB-32O4.2 |
| chr20_-_34638841 | 0.17 |
ENST00000565493.1
|
LINC00657
|
long intergenic non-protein coding RNA 657 |
| chr3_-_114035026 | 0.17 |
ENST00000570269.1
|
RP11-553L6.5
|
RP11-553L6.5 |
| chr2_+_85661918 | 0.17 |
ENST00000340326.2
|
SH2D6
|
SH2 domain containing 6 |
| chr18_-_55288973 | 0.17 |
ENST00000423481.2
ENST00000587194.1 ENST00000591599.1 ENST00000588661.1 |
NARS
|
asparaginyl-tRNA synthetase |
| chr2_+_138721850 | 0.17 |
ENST00000329366.4
ENST00000280097.3 |
HNMT
|
histamine N-methyltransferase |
| chr2_+_44502630 | 0.17 |
ENST00000410056.3
ENST00000409741.1 ENST00000409229.3 |
SLC3A1
|
solute carrier family 3 (amino acid transporter heavy chain), member 1 |
| chr2_+_44502597 | 0.17 |
ENST00000260649.6
ENST00000409387.1 |
SLC3A1
|
solute carrier family 3 (amino acid transporter heavy chain), member 1 |
| chr1_+_179335101 | 0.17 |
ENST00000508285.1
ENST00000511889.1 |
AXDND1
|
axonemal dynein light chain domain containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of BPTF
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 1.0 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.1 | 0.7 | GO:0045918 | negative regulation of cytolysis(GO:0045918) |
| 0.1 | 0.2 | GO:0090199 | regulation of release of cytochrome c from mitochondria(GO:0090199) |
| 0.1 | 0.3 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.6 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.1 | 0.3 | GO:0042245 | RNA repair(GO:0042245) |
| 0.1 | 0.4 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.1 | 0.3 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.1 | 0.7 | GO:0010133 | proline catabolic process(GO:0006562) proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 0.4 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.1 | 0.4 | GO:0097112 | gamma-aminobutyric acid receptor clustering(GO:0097112) |
| 0.1 | 0.3 | GO:0030070 | insulin processing(GO:0030070) |
| 0.1 | 0.2 | GO:0002590 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 0.1 | 0.4 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.1 | 0.2 | GO:0045720 | negative regulation of integrin biosynthetic process(GO:0045720) |
| 0.1 | 0.3 | GO:0051598 | meiotic recombination checkpoint(GO:0051598) |
| 0.1 | 0.2 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.1 | 0.2 | GO:0006147 | guanine catabolic process(GO:0006147) |
| 0.1 | 0.3 | GO:1904647 | response to rotenone(GO:1904647) |
| 0.1 | 0.3 | GO:0042231 | interleukin-13 biosynthetic process(GO:0042231) |
| 0.1 | 0.5 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.1 | 0.3 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.1 | 0.4 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.3 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.1 | 0.2 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.1 | 0.2 | GO:0046356 | acetyl-CoA catabolic process(GO:0046356) |
| 0.1 | 0.2 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.1 | 0.2 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.1 | 0.2 | GO:0070898 | RNA polymerase III transcriptional preinitiation complex assembly(GO:0070898) |
| 0.1 | 0.3 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.1 | 0.3 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.1 | 0.1 | GO:0007266 | Rho protein signal transduction(GO:0007266) |
| 0.1 | 0.4 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.1 | 0.3 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 0.2 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 | 0.2 | GO:0042495 | detection of triacyl bacterial lipopeptide(GO:0042495) detection of bacterial lipopeptide(GO:0070340) |
| 0.1 | 0.3 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.1 | 0.2 | GO:0035880 | embryonic nail plate morphogenesis(GO:0035880) |
| 0.1 | 0.2 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.1 | 0.2 | GO:2000360 | negative regulation of fertilization(GO:0060467) positive regulation of female gonad development(GO:2000196) negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.1 | 0.2 | GO:0034395 | regulation of transcription from RNA polymerase II promoter in response to iron(GO:0034395) |
| 0.1 | 0.4 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.1 | 0.1 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.1 | 0.2 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.1 | 0.3 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 0.2 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.0 | 0.1 | GO:0003250 | cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) |
| 0.0 | 0.2 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 0.0 | 0.3 | GO:2000661 | positive regulation of interleukin-1-mediated signaling pathway(GO:2000661) |
| 0.0 | 0.2 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 0.1 | GO:0071283 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) response to DDT(GO:0046680) cellular response to iron(III) ion(GO:0071283) histone H3-S10 phosphorylation involved in chromosome condensation(GO:2000775) |
| 0.0 | 0.2 | GO:1990022 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.0 | 0.1 | GO:0061110 | dense core granule biogenesis(GO:0061110) regulation of dense core granule biogenesis(GO:2000705) |
| 0.0 | 0.2 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.0 | 0.6 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.1 | GO:0034505 | tooth mineralization(GO:0034505) enamel mineralization(GO:0070166) |
| 0.0 | 0.9 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.2 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 0.2 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.0 | 0.4 | GO:0021943 | formation of radial glial scaffolds(GO:0021943) |
| 0.0 | 0.2 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 0.1 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.0 | 0.3 | GO:1902731 | negative regulation of chondrocyte proliferation(GO:1902731) |
| 0.0 | 0.7 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.2 | GO:1905244 | regulation of modification of synaptic structure(GO:1905244) |
| 0.0 | 0.4 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.0 | 0.2 | GO:0019285 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.0 | 0.1 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.1 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.0 | 0.3 | GO:2000490 | negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.0 | 0.1 | GO:0006272 | leading strand elongation(GO:0006272) |
| 0.0 | 0.2 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.6 | GO:2000194 | regulation of female gonad development(GO:2000194) |
| 0.0 | 1.0 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.2 | GO:1901073 | N-acetylglucosamine biosynthetic process(GO:0006045) glucosamine-containing compound biosynthetic process(GO:1901073) |
| 0.0 | 0.1 | GO:0043095 | regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 0.0 | 0.2 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:1901291 | negative regulation of double-strand break repair via single-strand annealing(GO:1901291) |
| 0.0 | 0.0 | GO:0042226 | interleukin-6 biosynthetic process(GO:0042226) |
| 0.0 | 0.2 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 0.1 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.3 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.1 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.0 | 0.4 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.3 | GO:0061517 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) |
| 0.0 | 0.2 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.0 | 0.2 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.0 | 0.1 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) double-strand break repair involved in meiotic recombination(GO:1990918) |
| 0.0 | 0.3 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.2 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.1 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0000915 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.3 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.5 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.1 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.0 | 0.1 | GO:0033499 | galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.0 | 0.1 | GO:0000717 | nucleotide-excision repair, DNA duplex unwinding(GO:0000717) |
| 0.0 | 0.1 | GO:0048597 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.0 | 0.2 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.0 | 0.4 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:0009304 | tRNA transcription(GO:0009304) |
| 0.0 | 0.2 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) |
| 0.0 | 1.2 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.4 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.0 | 0.4 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.0 | 0.1 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.3 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.0 | 0.2 | GO:0060809 | mesodermal to mesenchymal transition involved in gastrulation(GO:0060809) |
| 0.0 | 0.2 | GO:1901159 | glucuronate catabolic process(GO:0006064) glucuronate catabolic process to xylulose 5-phosphate(GO:0019640) xylulose 5-phosphate metabolic process(GO:0051167) xylulose 5-phosphate biosynthetic process(GO:1901159) |
| 0.0 | 0.2 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.1 | GO:2001247 | positive regulation of phosphatidylcholine biosynthetic process(GO:2001247) |
| 0.0 | 0.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.1 | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.0 | 0.1 | GO:1990569 | UDP-N-acetylglucosamine transport(GO:0015788) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.0 | 0.1 | GO:0015864 | uridine transport(GO:0015862) pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.3 | GO:0042262 | DNA protection(GO:0042262) |
| 0.0 | 0.1 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.0 | 0.3 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.1 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:1990654 | sebum secreting cell proliferation(GO:1990654) |
| 0.0 | 0.5 | GO:0061339 | establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.0 | 0.0 | GO:0045666 | positive regulation of neuron differentiation(GO:0045666) |
| 0.0 | 0.4 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.1 | GO:1903033 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.0 | 0.3 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.1 | GO:1904117 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.0 | 0.1 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 | 0.1 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.0 | 0.3 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.1 | GO:0046709 | IDP metabolic process(GO:0046707) IDP catabolic process(GO:0046709) |
| 0.0 | 0.1 | GO:0043126 | regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043126) positive regulation of 1-phosphatidylinositol 4-kinase activity(GO:0043128) |
| 0.0 | 0.1 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) |
| 0.0 | 0.1 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.2 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:0060167 | regulation of adenosine receptor signaling pathway(GO:0060167) positive regulation of adenosine receptor signaling pathway(GO:0060168) |
| 0.0 | 0.2 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 0.0 | 0.8 | GO:0050427 | purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.1 | GO:0035669 | TRAM-dependent toll-like receptor signaling pathway(GO:0035668) TRAM-dependent toll-like receptor 4 signaling pathway(GO:0035669) |
| 0.0 | 0.1 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.2 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.2 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.1 | GO:0038178 | complement component C5a signaling pathway(GO:0038178) |
| 0.0 | 0.1 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.0 | 0.1 | GO:0000921 | septin ring assembly(GO:0000921) septin ring organization(GO:0031106) |
| 0.0 | 0.3 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.0 | 0.1 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.3 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.1 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.0 | 0.1 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.1 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.0 | 0.3 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.1 | GO:0050848 | regulation of calcium-mediated signaling(GO:0050848) |
| 0.0 | 0.1 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.0 | 0.1 | GO:0006288 | base-excision repair, DNA ligation(GO:0006288) regulation of DNA ligase activity(GO:1904875) |
| 0.0 | 0.1 | GO:0052056 | modulation by virus of host molecular function(GO:0039506) suppression by virus of host molecular function(GO:0039507) suppression by virus of host catalytic activity(GO:0039513) modulation by virus of host catalytic activity(GO:0039516) suppression by virus of host cysteine-type endopeptidase activity involved in apoptotic process(GO:0039650) negative regulation by symbiont of host catalytic activity(GO:0052053) negative regulation by symbiont of host molecular function(GO:0052056) modulation by symbiont of host catalytic activity(GO:0052148) |
| 0.0 | 0.4 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.1 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.1 | GO:0014005 | microglia differentiation(GO:0014004) microglia development(GO:0014005) cellular response to norepinephrine stimulus(GO:0071874) |
| 0.0 | 0.1 | GO:1903352 | L-ornithine transmembrane transport(GO:1903352) |
| 0.0 | 0.2 | GO:1902659 | regulation of glucose mediated signaling pathway(GO:1902659) |
| 0.0 | 0.1 | GO:1902616 | acyl carnitine transport(GO:0006844) acyl carnitine transmembrane transport(GO:1902616) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.1 | GO:2000224 | sesquiterpenoid metabolic process(GO:0006714) sesquiterpenoid catabolic process(GO:0016107) farnesol metabolic process(GO:0016487) farnesol catabolic process(GO:0016488) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.1 | GO:0010041 | response to iron(III) ion(GO:0010041) |
| 0.0 | 0.2 | GO:0018406 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.1 | GO:0010992 | ubiquitin homeostasis(GO:0010992) |
| 0.0 | 0.2 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.1 | GO:2001170 | negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.0 | 0.1 | GO:0033967 | box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.0 | 0.2 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.0 | GO:2000537 | regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.0 | 0.1 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.0 | 0.1 | GO:0061740 | protein targeting to lysosome involved in chaperone-mediated autophagy(GO:0061740) |
| 0.0 | 0.1 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.0 | 0.1 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.0 | 0.2 | GO:0050878 | regulation of body fluid levels(GO:0050878) |
| 0.0 | 0.0 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.1 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.0 | 0.1 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.0 | 0.1 | GO:0032354 | response to follicle-stimulating hormone(GO:0032354) |
| 0.0 | 0.1 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.0 | 0.2 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.1 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.1 | GO:0008343 | adult feeding behavior(GO:0008343) |
| 0.0 | 0.3 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.0 | 0.1 | GO:0061370 | testosterone biosynthetic process(GO:0061370) |
| 0.0 | 0.1 | GO:0006824 | cobalt ion transport(GO:0006824) |
| 0.0 | 0.1 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.0 | 0.0 | GO:0072738 | response to diamide(GO:0072737) cellular response to diamide(GO:0072738) |
| 0.0 | 0.1 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.1 | GO:1905097 | regulation of guanyl-nucleotide exchange factor activity(GO:1905097) |
| 0.0 | 0.1 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.0 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.0 | 0.0 | GO:0018013 | N-terminal peptidyl-alanine methylation(GO:0018011) N-terminal peptidyl-alanine trimethylation(GO:0018012) N-terminal peptidyl-glycine methylation(GO:0018013) N-terminal peptidyl-proline dimethylation(GO:0018016) peptidyl-alanine modification(GO:0018194) N-terminal peptidyl-proline methylation(GO:0035568) N-terminal peptidyl-serine methylation(GO:0035570) N-terminal peptidyl-serine dimethylation(GO:0035572) N-terminal peptidyl-serine trimethylation(GO:0035573) |
| 0.0 | 0.1 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.0 | 0.1 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.0 | 0.1 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.1 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.0 | 0.1 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) |
| 0.0 | 0.1 | GO:0070541 | response to platinum ion(GO:0070541) |
| 0.0 | 0.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.1 | GO:0061084 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.0 | 0.1 | GO:0050807 | regulation of synapse organization(GO:0050807) |
| 0.0 | 0.2 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.1 | GO:0044856 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.0 | 0.2 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.2 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.1 | GO:0007231 | osmosensory signaling pathway(GO:0007231) |
| 0.0 | 0.1 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.0 | 0.1 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.1 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.0 | GO:0097205 | glomerular filtration(GO:0003094) renal filtration(GO:0097205) |
| 0.0 | 0.1 | GO:0070383 | DNA cytosine deamination(GO:0070383) |
| 0.0 | 0.1 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.2 | GO:0090656 | t-circle formation(GO:0090656) |
| 0.0 | 0.1 | GO:0033088 | negative regulation of immature T cell proliferation(GO:0033087) negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 | 0.1 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.1 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.1 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 0.1 | GO:0019216 | regulation of lipid metabolic process(GO:0019216) |
| 0.0 | 0.1 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.2 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.1 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.2 | GO:0034654 | nucleobase-containing compound biosynthetic process(GO:0034654) |
| 0.0 | 0.1 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.0 | 0.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.4 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:0070458 | cellular detoxification of nitrogen compound(GO:0070458) |
| 0.0 | 0.0 | GO:0072720 | cellular response to mycotoxin(GO:0036146) response to dithiothreitol(GO:0072720) |
| 0.0 | 0.0 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.1 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.0 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.1 | GO:0044789 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.0 | 0.1 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.0 | 0.1 | GO:0070911 | global genome nucleotide-excision repair(GO:0070911) |
| 0.0 | 0.1 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.0 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.2 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.1 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.1 | GO:0023014 | signal transduction by protein phosphorylation(GO:0023014) |
| 0.0 | 0.1 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.3 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.0 | 0.2 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.0 | 0.0 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.0 | 0.0 | GO:2001162 | regulation of histone H3-K79 methylation(GO:2001160) positive regulation of histone H3-K79 methylation(GO:2001162) |
| 0.0 | 0.1 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.0 | GO:0086097 | regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 | 0.2 | GO:0038180 | nerve growth factor signaling pathway(GO:0038180) |
| 0.0 | 0.3 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 0.1 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 | 0.0 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.0 | 0.1 | GO:0090669 | telomerase RNA stabilization(GO:0090669) |
| 0.0 | 0.1 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.1 | GO:0048875 | chemical homeostasis within a tissue(GO:0048875) |
| 0.0 | 0.0 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 0.0 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.2 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.0 | GO:0045776 | negative regulation of blood pressure(GO:0045776) |
| 0.0 | 0.1 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 | 0.1 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.1 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.0 | 0.1 | GO:1990822 | basic amino acid transmembrane transport(GO:1990822) |
| 0.0 | 0.0 | GO:1905033 | positive regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903762) positive regulation of ventricular cardiac muscle cell action potential(GO:1903947) positive regulation of membrane repolarization during ventricular cardiac muscle cell action potential(GO:1905026) positive regulation of membrane repolarization during cardiac muscle cell action potential(GO:1905033) |
| 0.0 | 0.1 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.1 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.0 | 0.0 | GO:0001971 | negative regulation of activation of membrane attack complex(GO:0001971) |
| 0.0 | 0.1 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.0 | GO:0060151 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.0 | 0.0 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.1 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.2 | GO:0030855 | epithelial cell differentiation(GO:0030855) |
| 0.0 | 0.0 | GO:1901143 | insulin catabolic process(GO:1901143) |
| 0.0 | 0.2 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.1 | GO:2000371 | regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.0 | 0.1 | GO:0032328 | alanine transport(GO:0032328) |
| 0.0 | 0.2 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.1 | GO:0034093 | positive regulation of maintenance of sister chromatid cohesion(GO:0034093) positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) |
| 0.0 | 0.2 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0020003 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 0.2 | 0.6 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 0.4 | GO:0016935 | glycine-gated chloride channel complex(GO:0016935) |
| 0.1 | 0.3 | GO:0042022 | interleukin-12 receptor complex(GO:0042022) |
| 0.1 | 0.3 | GO:1990075 | kinesin II complex(GO:0016939) periciliary membrane compartment(GO:1990075) |
| 0.1 | 0.1 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.1 | 0.2 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.1 | 0.9 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.1 | 0.2 | GO:0009346 | citrate lyase complex(GO:0009346) |
| 0.1 | 0.2 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.1 | 0.3 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.1 | 0.2 | GO:0070993 | translation preinitiation complex(GO:0070993) |
| 0.1 | 0.2 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.1 | 0.3 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.1 | 0.2 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.3 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.2 | GO:0097362 | MCM8-MCM9 complex(GO:0097362) |
| 0.0 | 0.2 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.3 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.2 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.0 | 0.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.1 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.2 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.0 | 1.0 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.1 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.2 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.1 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.0 | 0.6 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.1 | GO:0002947 | tumor necrosis factor receptor superfamily complex(GO:0002947) |
| 0.0 | 0.1 | GO:0001534 | radial spoke(GO:0001534) |
| 0.0 | 0.1 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.0 | 0.2 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.2 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.1 | GO:0097636 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.0 | 0.1 | GO:0005712 | chiasma(GO:0005712) |
| 0.0 | 0.3 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.2 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 0.3 | GO:0090576 | RNA polymerase III transcription factor complex(GO:0090576) |
| 0.0 | 0.1 | GO:0075341 | host cell PML body(GO:0075341) |
| 0.0 | 0.1 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 0.0 | 0.8 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.2 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.4 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.4 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0034515 | proteasome storage granule(GO:0034515) |
| 0.0 | 0.2 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.2 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.3 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.1 | GO:0035101 | FACT complex(GO:0035101) |
| 0.0 | 0.4 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.3 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.1 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.1 | GO:0032302 | MutSbeta complex(GO:0032302) |
| 0.0 | 0.1 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.0 | 0.1 | GO:0036457 | keratohyalin granule(GO:0036457) |
| 0.0 | 0.4 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.7 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.2 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.2 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.1 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.2 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.2 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.1 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.1 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.0 | 0.3 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.4 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.1 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.0 | 0.1 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.0 | 0.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.2 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.4 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.2 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.2 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.4 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.1 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.2 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.2 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 0.1 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.2 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.0 | GO:0005595 | collagen type XII trimer(GO:0005595) |
| 0.0 | 0.2 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.0 | GO:0000811 | GINS complex(GO:0000811) |
| 0.0 | 0.1 | GO:0072588 | box H/ACA snoRNP complex(GO:0031429) box H/ACA RNP complex(GO:0072588) |
| 0.0 | 0.2 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.2 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.1 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.1 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.1 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.0 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.1 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 0.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.2 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.0 | 0.4 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.2 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.3 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.2 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.2 | 0.7 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 0.4 | GO:0008441 | 3'(2'),5'-bisphosphate nucleotidase activity(GO:0008441) |
| 0.1 | 0.9 | GO:0004376 | glycolipid mannosyltransferase activity(GO:0004376) |
| 0.1 | 0.8 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.3 | GO:0035248 | alpha-1,4-N-acetylgalactosaminyltransferase activity(GO:0035248) |
| 0.1 | 0.4 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 0.6 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 0.1 | 0.4 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 0.3 | GO:0016517 | interleukin-12 receptor activity(GO:0016517) |
| 0.1 | 0.3 | GO:0060001 | minus-end directed microfilament motor activity(GO:0060001) |
| 0.1 | 0.3 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.3 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.1 | 0.3 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.1 | 0.4 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.1 | 0.2 | GO:0005151 | interleukin-1, Type II receptor binding(GO:0005151) |
| 0.1 | 0.3 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.1 | 0.4 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.1 | 0.2 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 0.1 | 0.2 | GO:0008892 | guanine deaminase activity(GO:0008892) |
| 0.1 | 0.5 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.1 | 0.3 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.1 | 0.2 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.1 | 0.5 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.1 | 0.4 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.1 | 0.3 | GO:0010851 | cyclase regulator activity(GO:0010851) guanylate cyclase regulator activity(GO:0030249) |
| 0.1 | 0.2 | GO:0051908 | double-stranded DNA 5'-3' exodeoxyribonuclease activity(GO:0051908) |
| 0.1 | 0.4 | GO:0000995 | transcription factor activity, core RNA polymerase III binding(GO:0000995) |
| 0.1 | 0.2 | GO:0004796 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.1 | 0.2 | GO:0070119 | ciliary neurotrophic factor binding(GO:0070119) |
| 0.1 | 0.2 | GO:0003878 | ATP citrate synthase activity(GO:0003878) |
| 0.1 | 0.2 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.1 | GO:0001003 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) |
| 0.0 | 0.6 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.2 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 0.5 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.3 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.2 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.2 | GO:0008761 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) N-acylmannosamine kinase activity(GO:0009384) |
| 0.0 | 0.3 | GO:0051748 | UTP:glucose-1-phosphate uridylyltransferase activity(GO:0003983) UTP-monosaccharide-1-phosphate uridylyltransferase activity(GO:0051748) |
| 0.0 | 0.3 | GO:0047522 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.0 | 0.2 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.0 | 0.3 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.0 | 0.1 | GO:0048244 | phytanoyl-CoA dioxygenase activity(GO:0048244) |
| 0.0 | 0.1 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.0 | 0.6 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.0 | 0.4 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.3 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.3 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.2 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.1 | GO:0044549 | GTP cyclohydrolase binding(GO:0044549) |
| 0.0 | 0.1 | GO:0034039 | 8-oxo-7,8-dihydroguanine DNA N-glycosylase activity(GO:0034039) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.1 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.1 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 0.4 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.2 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.0 | 0.1 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.0 | 0.2 | GO:0030613 | oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) |
| 0.0 | 0.1 | GO:0003955 | NAD(P)H dehydrogenase (quinone) activity(GO:0003955) 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.0 | 0.2 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.0 | 0.1 | GO:0001602 | peptide YY receptor activity(GO:0001601) pancreatic polypeptide receptor activity(GO:0001602) |
| 0.0 | 0.2 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.2 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.1 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.3 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.3 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.5 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) |
| 0.0 | 0.4 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.2 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.1 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.0 | 0.0 | GO:0033677 | DNA/RNA helicase activity(GO:0033677) |
| 0.0 | 1.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.2 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.4 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.1 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.1 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.0 | 0.1 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.1 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.0 | 0.2 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.1 | GO:0004362 | glutathione-disulfide reductase activity(GO:0004362) |
| 0.0 | 0.1 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.1 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.1 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.0 | 0.1 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.0 | 0.2 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.1 | GO:0004325 | ferrochelatase activity(GO:0004325) |
| 0.0 | 0.3 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.1 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.6 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.1 | GO:0004917 | interleukin-7 receptor activity(GO:0004917) |
| 0.0 | 0.0 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.2 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 0.1 | GO:0003774 | motor activity(GO:0003774) |
| 0.0 | 0.1 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.1 | GO:0070736 | protein-glycine ligase activity, initiating(GO:0070736) |
| 0.0 | 0.1 | GO:0086057 | voltage-gated calcium channel activity involved in bundle of His cell action potential(GO:0086057) |
| 0.0 | 0.1 | GO:0004878 | complement component C5a receptor activity(GO:0004878) |
| 0.0 | 0.4 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.0 | 0.1 | GO:0004057 | arginyltransferase activity(GO:0004057) |
| 0.0 | 0.1 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.1 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.3 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.1 | GO:0032356 | oxidized DNA binding(GO:0032356) |
| 0.0 | 0.2 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.0 | 0.1 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.1 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.0 | 0.1 | GO:0035034 | histone acetyltransferase regulator activity(GO:0035034) |
| 0.0 | 0.1 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.0 | 0.2 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.0 | 0.1 | GO:0032558 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.0 | 0.1 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.1 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.0 | 0.1 | GO:1990259 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.1 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.1 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.1 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.2 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.0 | 0.0 | GO:0015633 | zinc transporting ATPase activity(GO:0015633) |
| 0.0 | 0.1 | GO:0004492 | methylmalonyl-CoA decarboxylase activity(GO:0004492) |
| 0.0 | 0.2 | GO:0043199 | sulfate binding(GO:0043199) |
| 0.0 | 0.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.1 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.0 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.0 | 0.1 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.1 | GO:0052642 | lysophosphatidic acid phosphatase activity(GO:0052642) |
| 0.0 | 0.0 | GO:0046906 | tetrapyrrole binding(GO:0046906) |
| 0.0 | 0.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.0 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.0 | 0.0 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.1 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.0 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) |
| 0.0 | 0.1 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.0 | GO:0031626 | beta-endorphin binding(GO:0031626) |
| 0.0 | 0.0 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.0 | 0.1 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.2 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.1 | GO:0016005 | phospholipase A2 activator activity(GO:0016005) |
| 0.0 | 0.0 | GO:0080130 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.1 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.0 | 0.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.0 | 0.2 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.1 | GO:0019144 | ADP-sugar diphosphatase activity(GO:0019144) |
| 0.0 | 0.0 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.0 | 0.4 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.1 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.0 | 0.1 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.0 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 0.0 | 0.1 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.0 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.5 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.2 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.0 | GO:0005415 | nucleoside:sodium symporter activity(GO:0005415) |
| 0.0 | 0.0 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 0.0 | 0.2 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.1 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.3 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.1 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.0 | 1.2 | GO:0008094 | DNA-dependent ATPase activity(GO:0008094) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.0 | 0.1 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.0 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.0 | 0.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.0 | GO:0004803 | transposase activity(GO:0004803) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.1 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.2 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.2 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.2 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.2 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.5 | GO:0005109 | frizzled binding(GO:0005109) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 1.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.0 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.8 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 1.3 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.2 | ST STAT3 PATHWAY | STAT3 Pathway |
| 0.0 | 0.1 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 1.0 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.6 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.7 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.5 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.9 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.8 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.7 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.3 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.1 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF MITOTIC PROTEINS | Genes involved in APC/C:Cdc20 mediated degradation of mitotic proteins |
| 0.0 | 0.1 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 1.3 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.0 | 1.0 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.2 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.5 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.9 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.6 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 1.0 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.1 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.5 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 1.0 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.1 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.0 | 1.4 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.2 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.3 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.1 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.3 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.3 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.1 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.6 | REACTOME G2 M CHECKPOINTS | Genes involved in G2/M Checkpoints |
| 0.0 | 0.6 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.4 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.6 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.1 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.3 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.2 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.1 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.2 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.2 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.3 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |