Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
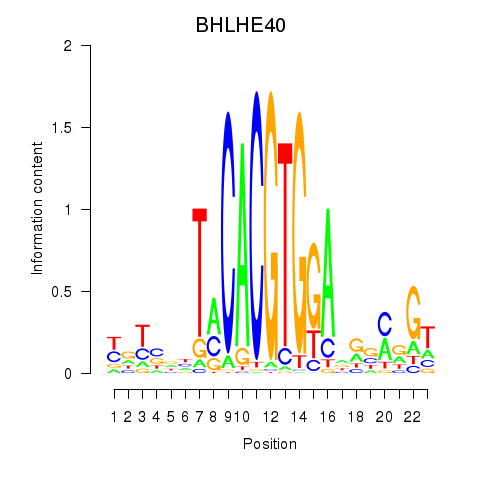
Results for BHLHE40
Z-value: 0.95
Transcription factors associated with BHLHE40
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BHLHE40
|
ENSG00000134107.4 | basic helix-loop-helix family member e40 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BHLHE40 | hg19_v2_chr3_+_5020801_5020952 | -0.65 | 3.5e-01 | Click! |
Activity profile of BHLHE40 motif
Sorted Z-values of BHLHE40 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_48688357 | 1.11 |
ENST00000533824.1
ENST00000438567.2 ENST00000236495.5 ENST00000420136.2 |
SLC5A9
|
solute carrier family 5 (sodium/sugar cotransporter), member 9 |
| chrX_+_23685653 | 0.93 |
ENST00000379331.3
|
PRDX4
|
peroxiredoxin 4 |
| chr19_+_54058073 | 0.70 |
ENST00000505949.1
ENST00000513265.1 |
ZNF331
|
zinc finger protein 331 |
| chrX_+_54556633 | 0.69 |
ENST00000336470.4
ENST00000360845.2 |
GNL3L
|
guanine nucleotide binding protein-like 3 (nucleolar)-like |
| chr4_+_119200215 | 0.67 |
ENST00000602573.1
|
SNHG8
|
small nucleolar RNA host gene 8 (non-protein coding) |
| chr13_+_50656307 | 0.58 |
ENST00000378180.4
|
DLEU1
|
deleted in lymphocytic leukemia 1 (non-protein coding) |
| chr8_-_99129338 | 0.58 |
ENST00000520507.1
|
HRSP12
|
heat-responsive protein 12 |
| chr16_-_19729453 | 0.55 |
ENST00000564480.1
|
KNOP1
|
lysine-rich nucleolar protein 1 |
| chrX_+_43515467 | 0.52 |
ENST00000338702.3
ENST00000542639.1 |
MAOA
|
monoamine oxidase A |
| chr6_-_151773232 | 0.52 |
ENST00000444024.1
ENST00000367303.4 |
RMND1
|
required for meiotic nuclear division 1 homolog (S. cerevisiae) |
| chr17_-_17109579 | 0.51 |
ENST00000321560.3
|
PLD6
|
phospholipase D family, member 6 |
| chr4_-_76861392 | 0.49 |
ENST00000505594.1
|
NAAA
|
N-acylethanolamine acid amidase |
| chr2_-_190649023 | 0.46 |
ENST00000409519.1
ENST00000458355.1 |
ORMDL1
|
ORM1-like 1 (S. cerevisiae) |
| chr4_-_15683118 | 0.43 |
ENST00000507899.1
ENST00000510802.1 |
FBXL5
|
F-box and leucine-rich repeat protein 5 |
| chr14_-_45722605 | 0.42 |
ENST00000310806.4
|
MIS18BP1
|
MIS18 binding protein 1 |
| chr8_-_99129384 | 0.41 |
ENST00000521560.1
ENST00000254878.3 |
HRSP12
|
heat-responsive protein 12 |
| chr19_+_45909893 | 0.39 |
ENST00000592852.1
ENST00000589804.1 ENST00000590794.1 |
CD3EAP
|
CD3e molecule, epsilon associated protein |
| chr13_+_111972980 | 0.39 |
ENST00000283547.1
|
TEX29
|
testis expressed 29 |
| chr13_+_35516390 | 0.38 |
ENST00000540320.1
ENST00000400445.3 ENST00000310336.4 |
NBEA
|
neurobeachin |
| chr9_+_131902283 | 0.38 |
ENST00000436883.1
ENST00000414510.1 |
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr4_+_119199904 | 0.37 |
ENST00000602483.1
ENST00000602819.1 |
SNHG8
|
small nucleolar RNA host gene 8 (non-protein coding) |
| chr6_-_36842784 | 0.36 |
ENST00000373699.5
|
PPIL1
|
peptidylprolyl isomerase (cyclophilin)-like 1 |
| chr4_-_159094194 | 0.34 |
ENST00000592057.1
ENST00000585682.1 ENST00000393807.5 |
FAM198B
|
family with sequence similarity 198, member B |
| chr8_+_109455830 | 0.34 |
ENST00000524143.1
|
EMC2
|
ER membrane protein complex subunit 2 |
| chrX_+_23685563 | 0.34 |
ENST00000379341.4
|
PRDX4
|
peroxiredoxin 4 |
| chr12_+_65004292 | 0.34 |
ENST00000542104.1
ENST00000336061.2 |
RASSF3
|
Ras association (RalGDS/AF-6) domain family member 3 |
| chr6_-_136871957 | 0.33 |
ENST00000354570.3
|
MAP7
|
microtubule-associated protein 7 |
| chr1_+_203830703 | 0.33 |
ENST00000414487.2
|
SNRPE
|
small nuclear ribonucleoprotein polypeptide E |
| chr7_-_37024665 | 0.31 |
ENST00000396040.2
|
ELMO1
|
engulfment and cell motility 1 |
| chr8_+_125985531 | 0.30 |
ENST00000319286.5
|
ZNF572
|
zinc finger protein 572 |
| chr8_-_134584092 | 0.28 |
ENST00000522652.1
|
ST3GAL1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr2_+_103236004 | 0.28 |
ENST00000233969.2
|
SLC9A2
|
solute carrier family 9, subfamily A (NHE2, cation proton antiporter 2), member 2 |
| chr10_+_101491968 | 0.27 |
ENST00000370476.5
ENST00000370472.4 |
CUTC
|
cutC copper transporter |
| chrX_+_66764375 | 0.26 |
ENST00000374690.3
|
AR
|
androgen receptor |
| chr11_-_57282349 | 0.26 |
ENST00000528450.1
|
SLC43A1
|
solute carrier family 43 (amino acid system L transporter), member 1 |
| chr2_+_183989157 | 0.26 |
ENST00000541912.1
|
NUP35
|
nucleoporin 35kDa |
| chr17_-_53046058 | 0.26 |
ENST00000571584.1
ENST00000299335.3 |
COX11
|
cytochrome c oxidase assembly homolog 11 (yeast) |
| chr10_+_49514698 | 0.25 |
ENST00000432379.1
ENST00000429041.1 ENST00000374189.1 |
MAPK8
|
mitogen-activated protein kinase 8 |
| chr2_+_190649107 | 0.25 |
ENST00000441310.2
ENST00000409985.1 ENST00000446877.1 ENST00000418224.3 ENST00000409823.3 ENST00000374826.4 ENST00000424766.1 ENST00000447232.2 ENST00000432292.3 |
PMS1
|
PMS1 postmeiotic segregation increased 1 (S. cerevisiae) |
| chr19_+_56154913 | 0.25 |
ENST00000270451.5
ENST00000588537.1 |
ZNF581
|
zinc finger protein 581 |
| chr17_+_7982800 | 0.24 |
ENST00000399413.3
|
AC129492.6
|
AC129492.6 |
| chr22_-_43411106 | 0.24 |
ENST00000453643.1
ENST00000263246.3 ENST00000337959.4 |
PACSIN2
|
protein kinase C and casein kinase substrate in neurons 2 |
| chr5_+_94982435 | 0.24 |
ENST00000511684.1
ENST00000380005.4 |
RFESD
|
Rieske (Fe-S) domain containing |
| chr14_-_45722360 | 0.23 |
ENST00000451174.1
|
MIS18BP1
|
MIS18 binding protein 1 |
| chr3_+_99536663 | 0.23 |
ENST00000421999.2
ENST00000463526.1 |
CMSS1
|
cms1 ribosomal small subunit homolog (yeast) |
| chr2_-_241497390 | 0.22 |
ENST00000272972.3
ENST00000401804.1 ENST00000361678.4 ENST00000405523.3 |
ANKMY1
|
ankyrin repeat and MYND domain containing 1 |
| chr21_+_27107672 | 0.22 |
ENST00000400075.3
|
GABPA
|
GA binding protein transcription factor, alpha subunit 60kDa |
| chr3_-_98312548 | 0.22 |
ENST00000264193.2
|
CPOX
|
coproporphyrinogen oxidase |
| chr3_+_133293278 | 0.22 |
ENST00000508481.1
ENST00000420115.2 ENST00000504867.1 ENST00000507408.1 ENST00000511392.1 ENST00000515421.1 |
CDV3
|
CDV3 homolog (mouse) |
| chr1_-_86174065 | 0.21 |
ENST00000370574.3
ENST00000431532.2 |
ZNHIT6
|
zinc finger, HIT-type containing 6 |
| chr8_+_22857048 | 0.21 |
ENST00000251822.6
|
RHOBTB2
|
Rho-related BTB domain containing 2 |
| chr9_+_130469257 | 0.21 |
ENST00000373295.2
|
C9orf117
|
chromosome 9 open reading frame 117 |
| chr4_-_100009856 | 0.20 |
ENST00000296412.8
|
ADH5
|
alcohol dehydrogenase 5 (class III), chi polypeptide |
| chr2_-_136743436 | 0.20 |
ENST00000441323.1
ENST00000449218.1 |
DARS
|
aspartyl-tRNA synthetase |
| chr1_+_173793777 | 0.20 |
ENST00000239457.5
|
DARS2
|
aspartyl-tRNA synthetase 2, mitochondrial |
| chrX_-_16887963 | 0.20 |
ENST00000380084.4
|
RBBP7
|
retinoblastoma binding protein 7 |
| chr17_-_10600818 | 0.19 |
ENST00000577427.1
ENST00000255390.5 |
SCO1
|
SCO1 cytochrome c oxidase assembly protein |
| chr9_-_95640218 | 0.19 |
ENST00000395506.3
ENST00000375495.3 ENST00000332591.6 |
ZNF484
|
zinc finger protein 484 |
| chr7_-_960521 | 0.19 |
ENST00000437486.1
|
ADAP1
|
ArfGAP with dual PH domains 1 |
| chr6_+_36410762 | 0.19 |
ENST00000483557.1
ENST00000498267.1 ENST00000544295.1 ENST00000449081.2 ENST00000536244.1 ENST00000460983.1 |
KCTD20
|
potassium channel tetramerization domain containing 20 |
| chr4_+_119199864 | 0.18 |
ENST00000602414.1
ENST00000602520.1 |
SNHG8
|
small nucleolar RNA host gene 8 (non-protein coding) |
| chr8_+_99129513 | 0.18 |
ENST00000522319.1
ENST00000401707.2 |
POP1
|
processing of precursor 1, ribonuclease P/MRP subunit (S. cerevisiae) |
| chr2_-_179343226 | 0.18 |
ENST00000434643.2
|
FKBP7
|
FK506 binding protein 7 |
| chr7_-_229557 | 0.18 |
ENST00000514988.1
|
AC145676.2
|
Uncharacterized protein |
| chr9_+_131901710 | 0.18 |
ENST00000524946.2
|
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr5_+_421030 | 0.17 |
ENST00000506456.1
|
AHRR
|
aryl-hydrocarbon receptor repressor |
| chr13_+_100741269 | 0.17 |
ENST00000376286.4
ENST00000376279.3 ENST00000376285.1 |
PCCA
|
propionyl CoA carboxylase, alpha polypeptide |
| chr1_-_207095324 | 0.17 |
ENST00000530505.1
ENST00000367091.3 ENST00000442471.2 |
FAIM3
|
Fas apoptotic inhibitory molecule 3 |
| chr9_+_131901661 | 0.17 |
ENST00000423100.1
|
PPP2R4
|
protein phosphatase 2A activator, regulatory subunit 4 |
| chr8_+_26240414 | 0.17 |
ENST00000380629.2
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chr2_-_136743039 | 0.16 |
ENST00000537273.1
|
DARS
|
aspartyl-tRNA synthetase |
| chr11_+_34664014 | 0.16 |
ENST00000527935.1
|
EHF
|
ets homologous factor |
| chr6_-_5260963 | 0.16 |
ENST00000464010.1
ENST00000468929.1 ENST00000480566.1 |
LYRM4
|
LYR motif containing 4 |
| chr12_+_54674482 | 0.16 |
ENST00000547708.1
ENST00000340913.6 ENST00000551702.1 ENST00000330752.8 ENST00000547276.1 |
HNRNPA1
|
heterogeneous nuclear ribonucleoprotein A1 |
| chr2_-_241497374 | 0.16 |
ENST00000373318.2
ENST00000406958.1 ENST00000391987.1 ENST00000373320.4 |
ANKMY1
|
ankyrin repeat and MYND domain containing 1 |
| chr1_+_21835858 | 0.15 |
ENST00000539907.1
ENST00000540617.1 ENST00000374840.3 |
ALPL
|
alkaline phosphatase, liver/bone/kidney |
| chr8_-_48872686 | 0.15 |
ENST00000314191.2
ENST00000338368.3 |
PRKDC
|
protein kinase, DNA-activated, catalytic polypeptide |
| chr2_+_207630081 | 0.14 |
ENST00000236980.6
ENST00000418289.1 ENST00000402774.3 ENST00000403094.3 |
FASTKD2
|
FAST kinase domains 2 |
| chr18_+_55712915 | 0.14 |
ENST00000592846.1
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr5_-_71616043 | 0.14 |
ENST00000508863.2
ENST00000522095.1 ENST00000513900.1 ENST00000515404.1 ENST00000457646.4 ENST00000261413.5 |
MRPS27
|
mitochondrial ribosomal protein S27 |
| chr2_-_112642267 | 0.14 |
ENST00000341068.3
|
ANAPC1
|
anaphase promoting complex subunit 1 |
| chr19_-_695427 | 0.14 |
ENST00000329267.7
|
PRSS57
|
protease, serine, 57 |
| chr1_-_200589859 | 0.14 |
ENST00000367350.4
|
KIF14
|
kinesin family member 14 |
| chr16_-_79804394 | 0.13 |
ENST00000567993.1
|
RP11-345M22.2
|
RP11-345M22.2 |
| chr1_-_231376867 | 0.13 |
ENST00000366649.2
ENST00000318906.2 ENST00000366651.3 |
C1orf131
|
chromosome 1 open reading frame 131 |
| chr5_+_36152163 | 0.13 |
ENST00000274255.6
|
SKP2
|
S-phase kinase-associated protein 2, E3 ubiquitin protein ligase |
| chr22_+_48885379 | 0.13 |
ENST00000336769.5
|
FAM19A5
|
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5 |
| chr11_+_73019282 | 0.13 |
ENST00000263674.3
|
ARHGEF17
|
Rho guanine nucleotide exchange factor (GEF) 17 |
| chr8_+_55047763 | 0.13 |
ENST00000260102.4
ENST00000519831.1 |
MRPL15
|
mitochondrial ribosomal protein L15 |
| chr2_-_10587897 | 0.12 |
ENST00000405333.1
ENST00000443218.1 |
ODC1
|
ornithine decarboxylase 1 |
| chr1_+_44445549 | 0.12 |
ENST00000356836.6
|
B4GALT2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr10_-_97416400 | 0.11 |
ENST00000371224.2
ENST00000371221.3 |
ALDH18A1
|
aldehyde dehydrogenase 18 family, member A1 |
| chr12_-_58165870 | 0.11 |
ENST00000257848.7
|
METTL1
|
methyltransferase like 1 |
| chr17_-_7193711 | 0.11 |
ENST00000571464.1
|
YBX2
|
Y box binding protein 2 |
| chr6_+_5261225 | 0.11 |
ENST00000324331.6
|
FARS2
|
phenylalanyl-tRNA synthetase 2, mitochondrial |
| chr22_+_41697520 | 0.11 |
ENST00000352645.4
|
ZC3H7B
|
zinc finger CCCH-type containing 7B |
| chr17_+_61851504 | 0.11 |
ENST00000359353.5
ENST00000389924.2 |
DDX42
|
DEAD (Asp-Glu-Ala-Asp) box helicase 42 |
| chr19_+_41497178 | 0.10 |
ENST00000324071.4
|
CYP2B6
|
cytochrome P450, family 2, subfamily B, polypeptide 6 |
| chr2_-_10588630 | 0.09 |
ENST00000234111.4
|
ODC1
|
ornithine decarboxylase 1 |
| chr11_-_6624801 | 0.09 |
ENST00000534343.1
ENST00000254605.6 |
RRP8
|
ribosomal RNA processing 8, methyltransferase, homolog (yeast) |
| chr3_-_51533966 | 0.09 |
ENST00000504652.1
|
VPRBP
|
Vpr (HIV-1) binding protein |
| chr1_+_118472343 | 0.09 |
ENST00000369441.3
ENST00000349139.5 |
WDR3
|
WD repeat domain 3 |
| chr1_+_201708992 | 0.09 |
ENST00000367295.1
|
NAV1
|
neuron navigator 1 |
| chr8_+_109455845 | 0.08 |
ENST00000220853.3
|
EMC2
|
ER membrane protein complex subunit 2 |
| chr17_+_61851157 | 0.08 |
ENST00000578681.1
ENST00000583590.1 |
DDX42
|
DEAD (Asp-Glu-Ala-Asp) box helicase 42 |
| chr2_+_201676908 | 0.08 |
ENST00000409226.1
ENST00000452790.2 |
BZW1
|
basic leucine zipper and W2 domains 1 |
| chr14_+_101359265 | 0.08 |
ENST00000599197.1
|
AL117190.3
|
Esophagus cancer-related gene-2 interaction susceptibility protein; Uncharacterized protein |
| chr13_-_99630233 | 0.08 |
ENST00000376460.1
ENST00000442173.1 |
DOCK9
|
dedicator of cytokinesis 9 |
| chr17_-_8066843 | 0.08 |
ENST00000404970.3
|
VAMP2
|
vesicle-associated membrane protein 2 (synaptobrevin 2) |
| chr1_+_9648921 | 0.08 |
ENST00000377376.4
ENST00000340305.5 ENST00000340381.6 |
TMEM201
|
transmembrane protein 201 |
| chr6_-_109804412 | 0.07 |
ENST00000230122.3
|
ZBTB24
|
zinc finger and BTB domain containing 24 |
| chr20_+_23331373 | 0.07 |
ENST00000254998.2
|
NXT1
|
NTF2-like export factor 1 |
| chr2_-_47143160 | 0.07 |
ENST00000409800.1
ENST00000409218.1 |
MCFD2
|
multiple coagulation factor deficiency 2 |
| chrX_-_128977364 | 0.06 |
ENST00000371064.3
|
ZDHHC9
|
zinc finger, DHHC-type containing 9 |
| chr3_+_63638372 | 0.06 |
ENST00000496807.1
|
SNTN
|
sentan, cilia apical structure protein |
| chr6_+_44215603 | 0.06 |
ENST00000371554.1
|
HSP90AB1
|
heat shock protein 90kDa alpha (cytosolic), class B member 1 |
| chr6_+_13615554 | 0.06 |
ENST00000451315.2
|
NOL7
|
nucleolar protein 7, 27kDa |
| chr1_+_213123915 | 0.06 |
ENST00000366968.4
ENST00000490792.1 |
VASH2
|
vasohibin 2 |
| chr9_+_37120536 | 0.06 |
ENST00000336755.5
ENST00000534928.1 ENST00000322831.6 |
ZCCHC7
|
zinc finger, CCHC domain containing 7 |
| chr4_-_57301748 | 0.05 |
ENST00000264220.2
|
PPAT
|
phosphoribosyl pyrophosphate amidotransferase |
| chr21_-_45079341 | 0.05 |
ENST00000443485.1
ENST00000291560.2 |
HSF2BP
|
heat shock transcription factor 2 binding protein |
| chr16_+_29911864 | 0.05 |
ENST00000308748.5
|
ASPHD1
|
aspartate beta-hydroxylase domain containing 1 |
| chr17_-_72772425 | 0.05 |
ENST00000578822.1
|
NAT9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr5_-_89770582 | 0.05 |
ENST00000316610.6
|
MBLAC2
|
metallo-beta-lactamase domain containing 2 |
| chr22_-_37505449 | 0.05 |
ENST00000406725.1
|
TMPRSS6
|
transmembrane protease, serine 6 |
| chr5_+_149737202 | 0.05 |
ENST00000451292.1
ENST00000377797.3 ENST00000445265.2 ENST00000323668.7 ENST00000439160.2 ENST00000394269.3 ENST00000427724.2 ENST00000504761.2 ENST00000513346.1 ENST00000515516.1 |
TCOF1
|
Treacher Collins-Franceschetti syndrome 1 |
| chr2_+_183989083 | 0.04 |
ENST00000295119.4
|
NUP35
|
nucleoporin 35kDa |
| chr9_-_123691047 | 0.04 |
ENST00000373887.3
|
TRAF1
|
TNF receptor-associated factor 1 |
| chr2_+_105654441 | 0.04 |
ENST00000258455.3
|
MRPS9
|
mitochondrial ribosomal protein S9 |
| chr15_-_74988281 | 0.04 |
ENST00000566828.1
ENST00000563009.1 ENST00000568176.1 ENST00000566243.1 ENST00000566219.1 ENST00000426797.3 ENST00000566119.1 ENST00000315127.4 |
EDC3
|
enhancer of mRNA decapping 3 |
| chr5_-_36152031 | 0.03 |
ENST00000296603.4
|
LMBRD2
|
LMBR1 domain containing 2 |
| chr5_-_89770171 | 0.03 |
ENST00000514906.1
|
MBLAC2
|
metallo-beta-lactamase domain containing 2 |
| chr17_+_15635561 | 0.03 |
ENST00000584301.1
ENST00000580596.1 ENST00000464963.1 ENST00000437605.2 ENST00000579428.1 |
TBC1D26
|
TBC1 domain family, member 26 |
| chr12_+_93861264 | 0.03 |
ENST00000549982.1
ENST00000361630.2 |
MRPL42
|
mitochondrial ribosomal protein L42 |
| chr1_+_119957554 | 0.03 |
ENST00000543831.1
ENST00000433745.1 ENST00000369416.3 |
HSD3B2
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 |
| chr1_+_43637996 | 0.03 |
ENST00000528956.1
ENST00000529956.1 |
WDR65
|
WD repeat domain 65 |
| chr20_-_42939782 | 0.02 |
ENST00000396825.3
|
FITM2
|
fat storage-inducing transmembrane protein 2 |
| chr10_-_35379524 | 0.02 |
ENST00000374751.3
ENST00000374742.1 ENST00000602371.1 |
CUL2
|
cullin 2 |
| chrX_-_47518498 | 0.02 |
ENST00000335890.2
|
UXT
|
ubiquitously-expressed, prefoldin-like chaperone |
| chr2_-_179343268 | 0.02 |
ENST00000424785.2
|
FKBP7
|
FK506 binding protein 7 |
| chr14_+_73393040 | 0.02 |
ENST00000358377.2
ENST00000353777.3 ENST00000394234.2 ENST00000509153.1 ENST00000555042.1 |
DCAF4
|
DDB1 and CUL4 associated factor 4 |
| chr19_-_45953983 | 0.02 |
ENST00000592083.1
|
ERCC1
|
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence) |
| chr19_-_55652290 | 0.02 |
ENST00000589745.1
|
TNNT1
|
troponin T type 1 (skeletal, slow) |
| chr1_+_213123976 | 0.01 |
ENST00000366965.2
ENST00000366967.2 |
VASH2
|
vasohibin 2 |
| chr16_+_29911666 | 0.01 |
ENST00000563177.1
ENST00000483405.1 |
ASPHD1
|
aspartate beta-hydroxylase domain containing 1 |
| chr12_-_54673871 | 0.01 |
ENST00000209875.4
|
CBX5
|
chromobox homolog 5 |
| chr2_-_47142884 | 0.00 |
ENST00000409105.1
ENST00000409973.1 ENST00000409913.1 ENST00000319466.4 |
MCFD2
|
multiple coagulation factor deficiency 2 |
| chr22_-_38245304 | 0.00 |
ENST00000609454.1
|
ANKRD54
|
ankyrin repeat domain 54 |
| chr17_-_72772462 | 0.00 |
ENST00000582870.1
ENST00000581136.1 ENST00000357814.3 ENST00000579218.1 ENST00000583476.1 ENST00000580301.1 ENST00000583757.1 ENST00000582524.1 |
NAT9
|
N-acetyltransferase 9 (GCN5-related, putative) |
| chr14_+_77924373 | 0.00 |
ENST00000216479.3
ENST00000535854.2 ENST00000555517.1 |
AHSA1
|
AHA1, activator of heat shock 90kDa protein ATPase homolog 1 (yeast) |
| chr5_+_6766004 | 0.00 |
ENST00000506093.1
|
RP11-332J15.3
|
RP11-332J15.3 |
| chr19_+_7660716 | 0.00 |
ENST00000160298.4
ENST00000446248.2 |
CAMSAP3
|
calmodulin regulated spectrin-associated protein family, member 3 |
| chr20_-_35374456 | 0.00 |
ENST00000373803.2
ENST00000359675.2 ENST00000540765.1 ENST00000349004.1 |
NDRG3
|
NDRG family member 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of BHLHE40
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.1 | 0.5 | GO:0010636 | positive regulation of mitochondrial fusion(GO:0010636) |
| 0.1 | 0.3 | GO:0045720 | negative regulation of integrin biosynthetic process(GO:0045720) |
| 0.1 | 0.3 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.1 | 0.4 | GO:0090156 | cellular sphingolipid homeostasis(GO:0090156) |
| 0.1 | 0.3 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.1 | 0.7 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 | 0.2 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 0.1 | 0.2 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.1 | GO:0071529 | cementum mineralization(GO:0071529) |
| 0.0 | 0.5 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.0 | 0.3 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.2 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.2 | GO:0070370 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 0.5 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.2 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.1 | GO:1902775 | mitochondrial ribosome assembly(GO:0061668) mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.1 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.0 | 0.2 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.0 | 0.2 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.2 | GO:1903936 | response to sodium arsenite(GO:1903935) cellular response to sodium arsenite(GO:1903936) |
| 0.0 | 0.2 | GO:0016078 | tRNA catabolic process(GO:0016078) |
| 0.0 | 1.1 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.1 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.1 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.0 | 0.2 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.0 | 0.1 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.2 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.0 | 0.1 | GO:0006591 | ornithine metabolic process(GO:0006591) |
| 0.0 | 0.1 | GO:0002326 | B cell lineage commitment(GO:0002326) immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.4 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.7 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.2 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.3 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.4 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.3 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.7 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 0.2 | GO:0034753 | nuclear aryl hydrocarbon receptor complex(GO:0034753) |
| 0.0 | 0.3 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.1 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 0.4 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.2 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.2 | GO:0030430 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.0 | 0.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 1.3 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.7 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.2 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.1 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.3 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 0.3 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.2 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 1.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 0.5 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.1 | 0.3 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.1 | 0.5 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 0.2 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.0 | 0.3 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.0 | 0.3 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.3 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.2 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.2 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.2 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0002134 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.2 | GO:0004075 | biotin carboxylase activity(GO:0004075) biotin binding(GO:0009374) |
| 0.0 | 0.2 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.3 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.0 | 0.9 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |