Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
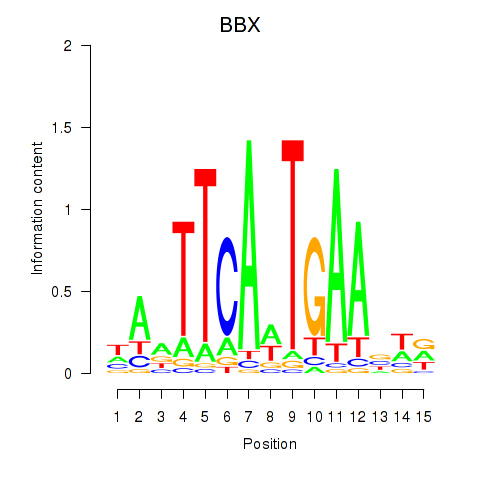
Results for BBX
Z-value: 0.81
Transcription factors associated with BBX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BBX
|
ENSG00000114439.14 | BBX high mobility group box domain containing |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BBX | hg19_v2_chr3_+_107364769_107364793 | -0.45 | 5.5e-01 | Click! |
Activity profile of BBX motif
Sorted Z-values of BBX motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_155484103 | 1.36 |
ENST00000302068.4
|
FGB
|
fibrinogen beta chain |
| chr4_+_155484155 | 1.32 |
ENST00000509493.1
|
FGB
|
fibrinogen beta chain |
| chr9_-_116837249 | 0.89 |
ENST00000466610.2
|
AMBP
|
alpha-1-microglobulin/bikunin precursor |
| chr12_-_10601963 | 0.69 |
ENST00000543893.1
|
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1 |
| chr5_+_140514782 | 0.51 |
ENST00000231134.5
|
PCDHB5
|
protocadherin beta 5 |
| chr20_-_7921090 | 0.48 |
ENST00000378789.3
|
HAO1
|
hydroxyacid oxidase (glycolate oxidase) 1 |
| chr14_-_23292596 | 0.39 |
ENST00000554741.1
|
SLC7A7
|
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr2_+_197577841 | 0.37 |
ENST00000409270.1
|
CCDC150
|
coiled-coil domain containing 150 |
| chr6_-_107235331 | 0.36 |
ENST00000433965.1
ENST00000430094.1 |
RP1-60O19.1
|
RP1-60O19.1 |
| chr7_-_16872932 | 0.35 |
ENST00000419572.2
ENST00000412973.1 |
AGR2
|
anterior gradient 2 |
| chr11_+_18154059 | 0.34 |
ENST00000531264.1
|
MRGPRX3
|
MAS-related GPR, member X3 |
| chr1_+_229440129 | 0.33 |
ENST00000366688.3
|
SPHAR
|
S-phase response (cyclin related) |
| chr19_+_47421933 | 0.30 |
ENST00000404338.3
|
ARHGAP35
|
Rho GTPase activating protein 35 |
| chr4_-_104021009 | 0.30 |
ENST00000509245.1
ENST00000296424.4 |
BDH2
|
3-hydroxybutyrate dehydrogenase, type 2 |
| chr6_-_107235287 | 0.30 |
ENST00000436659.1
ENST00000428750.1 ENST00000427903.1 |
RP1-60O19.1
|
RP1-60O19.1 |
| chr2_-_120124258 | 0.28 |
ENST00000409877.1
ENST00000409523.1 ENST00000409466.2 ENST00000414534.1 |
C2orf76
|
chromosome 2 open reading frame 76 |
| chr2_-_55237484 | 0.28 |
ENST00000394609.2
|
RTN4
|
reticulon 4 |
| chr7_-_16921601 | 0.28 |
ENST00000402239.3
ENST00000310398.2 ENST00000414935.1 |
AGR3
|
anterior gradient 3 |
| chr4_-_104020968 | 0.27 |
ENST00000504285.1
|
BDH2
|
3-hydroxybutyrate dehydrogenase, type 2 |
| chr3_+_51851612 | 0.26 |
ENST00000456080.1
|
IQCF3
|
IQ motif containing F3 |
| chr9_+_35161998 | 0.26 |
ENST00000396787.1
ENST00000378495.3 ENST00000378496.4 |
UNC13B
|
unc-13 homolog B (C. elegans) |
| chr7_+_135611542 | 0.25 |
ENST00000416501.1
|
AC015987.2
|
AC015987.2 |
| chr12_-_49581152 | 0.22 |
ENST00000550811.1
|
TUBA1A
|
tubulin, alpha 1a |
| chr6_-_25874440 | 0.22 |
ENST00000361703.6
ENST00000397060.4 |
SLC17A3
|
solute carrier family 17 (organic anion transporter), member 3 |
| chr12_-_39734783 | 0.21 |
ENST00000552961.1
|
KIF21A
|
kinesin family member 21A |
| chr1_+_95616933 | 0.21 |
ENST00000604203.1
|
RP11-57H12.6
|
TMEM56-RWDD3 readthrough |
| chr5_+_140729649 | 0.20 |
ENST00000523390.1
|
PCDHGB1
|
protocadherin gamma subfamily B, 1 |
| chr1_-_206785789 | 0.20 |
ENST00000437518.1
ENST00000367114.3 |
EIF2D
|
eukaryotic translation initiation factor 2D |
| chr7_-_130080977 | 0.20 |
ENST00000223208.5
|
CEP41
|
centrosomal protein 41kDa |
| chr5_+_74011328 | 0.19 |
ENST00000513336.1
|
HEXB
|
hexosaminidase B (beta polypeptide) |
| chr12_-_10978957 | 0.18 |
ENST00000240619.2
|
TAS2R10
|
taste receptor, type 2, member 10 |
| chr17_-_8263538 | 0.18 |
ENST00000535173.1
|
AC135178.1
|
HCG1985372; Uncharacterized protein; cDNA FLJ37541 fis, clone BRCAN2026340 |
| chr4_-_113558014 | 0.17 |
ENST00000503172.1
ENST00000505019.1 ENST00000309071.5 |
C4orf21
|
chromosome 4 open reading frame 21 |
| chr10_+_115674530 | 0.17 |
ENST00000451472.1
|
AL162407.1
|
CDNA FLJ20147 fis, clone COL07954; HCG1781466; Uncharacterized protein |
| chr9_-_95244781 | 0.16 |
ENST00000375544.3
ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN
|
asporin |
| chrX_+_100663243 | 0.16 |
ENST00000316594.5
|
HNRNPH2
|
heterogeneous nuclear ribonucleoprotein H2 (H') |
| chrX_-_119077695 | 0.16 |
ENST00000371410.3
|
NKAP
|
NFKB activating protein |
| chr4_-_987164 | 0.15 |
ENST00000398520.2
|
SLC26A1
|
solute carrier family 26 (anion exchanger), member 1 |
| chr3_+_81043000 | 0.15 |
ENST00000482617.1
|
RP11-6B4.1
|
RP11-6B4.1 |
| chr4_-_70080449 | 0.15 |
ENST00000446444.1
|
UGT2B11
|
UDP glucuronosyltransferase 2 family, polypeptide B11 |
| chr12_+_50690489 | 0.14 |
ENST00000598429.1
|
AC140061.12
|
Uncharacterized protein |
| chr15_+_49715293 | 0.13 |
ENST00000267843.4
ENST00000560270.1 |
FGF7
|
fibroblast growth factor 7 |
| chr1_+_170115142 | 0.13 |
ENST00000439373.2
|
METTL11B
|
methyltransferase like 11B |
| chr4_+_174818390 | 0.13 |
ENST00000509968.1
ENST00000512263.1 |
RP11-161D15.1
|
RP11-161D15.1 |
| chr1_+_212475148 | 0.13 |
ENST00000537030.3
|
PPP2R5A
|
protein phosphatase 2, regulatory subunit B', alpha |
| chr18_-_13915530 | 0.13 |
ENST00000327606.3
|
MC2R
|
melanocortin 2 receptor (adrenocorticotropic hormone) |
| chr14_+_75230011 | 0.12 |
ENST00000552421.1
ENST00000325680.7 ENST00000238571.3 |
YLPM1
|
YLP motif containing 1 |
| chr4_-_987217 | 0.11 |
ENST00000361661.2
ENST00000398516.2 |
SLC26A1
|
solute carrier family 26 (anion exchanger), member 1 |
| chr14_+_94492674 | 0.11 |
ENST00000203664.5
ENST00000553723.1 |
OTUB2
|
OTU domain, ubiquitin aldehyde binding 2 |
| chr22_-_39928823 | 0.11 |
ENST00000334678.3
|
RPS19BP1
|
ribosomal protein S19 binding protein 1 |
| chr6_-_107235378 | 0.11 |
ENST00000606430.1
|
RP1-60O19.1
|
RP1-60O19.1 |
| chr2_+_13677795 | 0.11 |
ENST00000434509.1
|
AC092635.1
|
AC092635.1 |
| chr1_+_55464600 | 0.10 |
ENST00000371265.4
|
BSND
|
Bartter syndrome, infantile, with sensorineural deafness (Barttin) |
| chr14_+_22458631 | 0.10 |
ENST00000390444.1
|
TRAV16
|
T cell receptor alpha variable 16 |
| chr2_+_102927962 | 0.09 |
ENST00000233954.1
ENST00000393393.3 ENST00000410040.1 |
IL1RL1
IL18R1
|
interleukin 1 receptor-like 1 interleukin 18 receptor 1 |
| chr16_+_53483983 | 0.09 |
ENST00000544545.1
|
RBL2
|
retinoblastoma-like 2 (p130) |
| chr9_-_5833027 | 0.09 |
ENST00000339450.5
|
ERMP1
|
endoplasmic reticulum metallopeptidase 1 |
| chr13_+_50570019 | 0.09 |
ENST00000442421.1
|
TRIM13
|
tripartite motif containing 13 |
| chr2_-_62081148 | 0.09 |
ENST00000404929.1
|
FAM161A
|
family with sequence similarity 161, member A |
| chr15_+_65823092 | 0.08 |
ENST00000566074.1
|
PTPLAD1
|
protein tyrosine phosphatase-like A domain containing 1 |
| chr3_-_47950745 | 0.08 |
ENST00000429422.1
|
MAP4
|
microtubule-associated protein 4 |
| chr2_+_63816295 | 0.07 |
ENST00000539945.1
ENST00000544381.1 |
MDH1
|
malate dehydrogenase 1, NAD (soluble) |
| chr3_-_133380731 | 0.07 |
ENST00000260810.5
|
TOPBP1
|
topoisomerase (DNA) II binding protein 1 |
| chr18_-_56985776 | 0.07 |
ENST00000587244.1
|
CPLX4
|
complexin 4 |
| chr2_-_62081254 | 0.07 |
ENST00000405894.3
|
FAM161A
|
family with sequence similarity 161, member A |
| chr3_+_127770455 | 0.07 |
ENST00000464451.1
|
SEC61A1
|
Sec61 alpha 1 subunit (S. cerevisiae) |
| chr9_-_95166884 | 0.07 |
ENST00000375561.5
|
OGN
|
osteoglycin |
| chr4_+_108910870 | 0.07 |
ENST00000403312.1
ENST00000603302.1 ENST00000309522.3 |
HADH
|
hydroxyacyl-CoA dehydrogenase |
| chr9_-_13175823 | 0.06 |
ENST00000545857.1
|
MPDZ
|
multiple PDZ domain protein |
| chr6_+_151358048 | 0.05 |
ENST00000450635.1
|
MTHFD1L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
| chr2_+_232135245 | 0.05 |
ENST00000446447.1
|
ARMC9
|
armadillo repeat containing 9 |
| chr9_-_95166841 | 0.05 |
ENST00000262551.4
|
OGN
|
osteoglycin |
| chr5_+_147774275 | 0.05 |
ENST00000513826.1
|
FBXO38
|
F-box protein 38 |
| chr6_-_150067696 | 0.05 |
ENST00000340413.2
ENST00000367403.3 |
NUP43
|
nucleoporin 43kDa |
| chr5_-_148929848 | 0.04 |
ENST00000504676.1
ENST00000515435.1 |
CSNK1A1
|
casein kinase 1, alpha 1 |
| chr21_+_19617140 | 0.04 |
ENST00000299295.2
ENST00000338326.3 |
CHODL
|
chondrolectin |
| chr14_+_38264418 | 0.04 |
ENST00000267368.7
ENST00000382320.3 |
TTC6
|
tetratricopeptide repeat domain 6 |
| chr1_+_117963209 | 0.04 |
ENST00000449370.2
|
MAN1A2
|
mannosidase, alpha, class 1A, member 2 |
| chr6_-_133084564 | 0.03 |
ENST00000532012.1
|
VNN2
|
vanin 2 |
| chr4_+_187813673 | 0.03 |
ENST00000606881.1
|
RP11-11N5.3
|
RP11-11N5.3 |
| chr7_+_128349106 | 0.03 |
ENST00000485070.1
|
FAM71F1
|
family with sequence similarity 71, member F1 |
| chr4_-_103998060 | 0.03 |
ENST00000339611.4
|
SLC9B2
|
solute carrier family 9, subfamily B (NHA2, cation proton antiporter 2), member 2 |
| chr18_-_21017817 | 0.03 |
ENST00000542162.1
ENST00000383233.3 ENST00000582336.1 ENST00000450466.2 ENST00000578520.1 ENST00000399707.1 |
TMEM241
|
transmembrane protein 241 |
| chr3_-_168865522 | 0.03 |
ENST00000464456.1
|
MECOM
|
MDS1 and EVI1 complex locus |
| chr6_+_31895287 | 0.03 |
ENST00000447952.2
|
C2
|
complement component 2 |
| chr4_-_103998439 | 0.02 |
ENST00000503230.1
ENST00000503818.1 |
SLC9B2
|
solute carrier family 9, subfamily B (NHA2, cation proton antiporter 2), member 2 |
| chr12_-_8765446 | 0.02 |
ENST00000537228.1
ENST00000229335.6 |
AICDA
|
activation-induced cytidine deaminase |
| chr7_+_95171457 | 0.02 |
ENST00000601424.1
|
AC002451.1
|
Protein LOC100996577 |
| chrX_+_144328348 | 0.02 |
ENST00000370493.3
|
SPANXN1
|
SPANX family, member N1 |
| chr14_-_81425828 | 0.01 |
ENST00000555529.1
ENST00000556042.1 ENST00000556981.1 |
CEP128
|
centrosomal protein 128kDa |
| chr13_+_33160553 | 0.01 |
ENST00000315596.10
|
PDS5B
|
PDS5, regulator of cohesion maintenance, homolog B (S. cerevisiae) |
| chr3_+_130569429 | 0.00 |
ENST00000505330.1
ENST00000504381.1 ENST00000507488.2 ENST00000393221.4 |
ATP2C1
|
ATPase, Ca++ transporting, type 2C, member 1 |
| chrX_+_54834159 | 0.00 |
ENST00000375053.2
ENST00000347546.4 ENST00000375062.4 |
MAGED2
|
melanoma antigen family D, 2 |
| chr6_-_133084580 | 0.00 |
ENST00000525270.1
ENST00000530536.1 ENST00000524919.1 |
VNN2
|
vanin 2 |
| chr16_+_56672571 | 0.00 |
ENST00000290705.8
|
MT1A
|
metallothionein 1A |
Network of associatons between targets according to the STRING database.
First level regulatory network of BBX
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.7 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.9 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.1 | 0.5 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.1 | 0.3 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 0.1 | 0.4 | GO:1903899 | lung goblet cell differentiation(GO:0060480) positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.0 | 0.3 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.2 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.0 | 0.4 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) basic amino acid transmembrane transport(GO:1990822) |
| 0.0 | 0.1 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.0 | 0.2 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.6 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.3 | GO:0050428 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.0 | 0.1 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.3 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.1 | GO:0090219 | negative regulation of lipid kinase activity(GO:0090219) |
| 0.0 | 0.2 | GO:0001731 | formation of translation preinitiation complex(GO:0001731) |
| 0.0 | 0.2 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.5 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.1 | GO:0009257 | 10-formyltetrahydrofolate biosynthetic process(GO:0009257) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.7 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.2 | GO:0044305 | calyx of Held(GO:0044305) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0019862 | IgA binding(GO:0019862) |
| 0.2 | 0.5 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.1 | 0.6 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.1 | 0.7 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.1 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.0 | 0.2 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.2 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.1 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.4 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.1 | GO:0047860 | diiodophenylpyruvate reductase activity(GO:0047860) |
| 0.0 | 0.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.5 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 2.7 | GO:0051087 | chaperone binding(GO:0051087) |
| 0.0 | 0.0 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.0 | 0.1 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.0 | 0.1 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.1 | GO:0004329 | formate-tetrahydrofolate ligase activity(GO:0004329) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.7 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.7 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |