Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
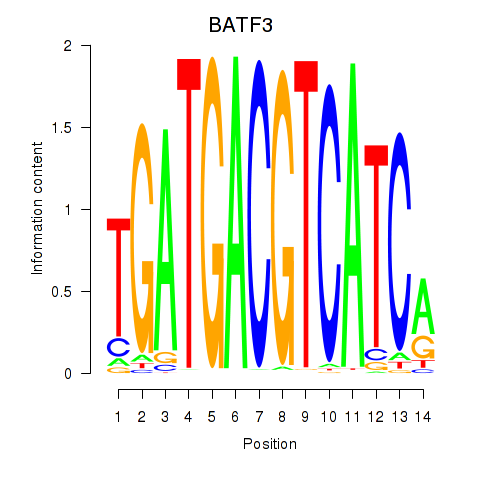
Results for BATF3
Z-value: 0.72
Transcription factors associated with BATF3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BATF3
|
ENSG00000123685.4 | basic leucine zipper ATF-like transcription factor 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| BATF3 | hg19_v2_chr1_-_212873267_212873332 | 0.83 | 1.7e-01 | Click! |
Activity profile of BATF3 motif
Sorted Z-values of BATF3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_77495007 | 0.94 |
ENST00000238647.3
|
IRF2BPL
|
interferon regulatory factor 2 binding protein-like |
| chr19_+_18723660 | 0.62 |
ENST00000262817.3
|
TMEM59L
|
transmembrane protein 59-like |
| chr22_+_38597889 | 0.55 |
ENST00000338483.2
ENST00000538320.1 ENST00000538999.1 ENST00000441709.1 |
MAFF
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog F |
| chr16_+_3068393 | 0.53 |
ENST00000573001.1
|
TNFRSF12A
|
tumor necrosis factor receptor superfamily, member 12A |
| chr3_-_119182506 | 0.45 |
ENST00000468676.1
|
TMEM39A
|
transmembrane protein 39A |
| chr10_+_35484793 | 0.43 |
ENST00000488741.1
ENST00000474931.1 ENST00000468236.1 ENST00000344351.5 ENST00000490511.1 |
CREM
|
cAMP responsive element modulator |
| chr17_-_48277552 | 0.40 |
ENST00000507689.1
|
COL1A1
|
collagen, type I, alpha 1 |
| chr2_-_202645612 | 0.36 |
ENST00000409632.2
ENST00000410052.1 ENST00000467448.1 |
ALS2
|
amyotrophic lateral sclerosis 2 (juvenile) |
| chr2_+_70142232 | 0.32 |
ENST00000540449.1
|
MXD1
|
MAX dimerization protein 1 |
| chr6_+_32939964 | 0.31 |
ENST00000607833.1
|
BRD2
|
bromodomain containing 2 |
| chr6_+_37400974 | 0.30 |
ENST00000455891.1
ENST00000373451.4 |
CMTR1
|
cap methyltransferase 1 |
| chr12_-_123380610 | 0.28 |
ENST00000535765.1
|
VPS37B
|
vacuolar protein sorting 37 homolog B (S. cerevisiae) |
| chr2_+_70142189 | 0.27 |
ENST00000264444.2
|
MXD1
|
MAX dimerization protein 1 |
| chr11_+_12696102 | 0.27 |
ENST00000527636.1
ENST00000527376.1 |
TEAD1
|
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
| chr19_+_8455077 | 0.27 |
ENST00000328024.6
|
RAB11B
|
RAB11B, member RAS oncogene family |
| chr10_-_128210005 | 0.25 |
ENST00000284694.7
ENST00000454341.1 ENST00000432642.1 ENST00000392694.1 |
C10orf90
|
chromosome 10 open reading frame 90 |
| chr3_-_119182453 | 0.24 |
ENST00000491685.1
ENST00000461654.1 |
TMEM39A
|
transmembrane protein 39A |
| chr6_+_31939608 | 0.24 |
ENST00000375331.2
ENST00000375333.2 |
STK19
|
serine/threonine kinase 19 |
| chr5_-_175965008 | 0.24 |
ENST00000537487.1
|
RNF44
|
ring finger protein 44 |
| chr3_-_52312636 | 0.22 |
ENST00000296490.3
|
WDR82
|
WD repeat domain 82 |
| chr14_-_50053081 | 0.22 |
ENST00000396020.3
ENST00000245458.6 |
RPS29
|
ribosomal protein S29 |
| chr12_+_7282795 | 0.22 |
ENST00000266546.6
|
CLSTN3
|
calsyntenin 3 |
| chr19_+_8455200 | 0.22 |
ENST00000601897.1
ENST00000594216.1 |
RAB11B
|
RAB11B, member RAS oncogene family |
| chr2_-_220264703 | 0.21 |
ENST00000519905.1
ENST00000523282.1 ENST00000434339.1 ENST00000457935.1 |
DNPEP
|
aspartyl aminopeptidase |
| chr6_-_91006461 | 0.21 |
ENST00000257749.4
ENST00000343122.3 ENST00000406998.2 ENST00000453877.1 |
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2 |
| chr2_+_220071490 | 0.20 |
ENST00000409206.1
ENST00000409594.1 ENST00000289528.5 ENST00000422255.1 ENST00000409412.1 ENST00000409097.1 ENST00000409336.1 ENST00000409217.1 ENST00000409319.1 ENST00000444522.2 |
ZFAND2B
|
zinc finger, AN1-type domain 2B |
| chr8_-_67090825 | 0.19 |
ENST00000276571.3
|
CRH
|
corticotropin releasing hormone |
| chr12_+_10365082 | 0.16 |
ENST00000545859.1
|
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr19_-_47616992 | 0.15 |
ENST00000253048.5
|
ZC3H4
|
zinc finger CCCH-type containing 4 |
| chr19_-_5340730 | 0.15 |
ENST00000372412.4
ENST00000357368.4 ENST00000262963.6 ENST00000348075.2 ENST00000353284.2 |
PTPRS
|
protein tyrosine phosphatase, receptor type, S |
| chr19_-_460996 | 0.15 |
ENST00000264554.6
|
SHC2
|
SHC (Src homology 2 domain containing) transforming protein 2 |
| chr2_+_74757050 | 0.15 |
ENST00000352222.3
ENST00000437202.1 |
HTRA2
|
HtrA serine peptidase 2 |
| chr11_-_65793948 | 0.15 |
ENST00000312106.5
|
CATSPER1
|
cation channel, sperm associated 1 |
| chrX_+_48398053 | 0.14 |
ENST00000537536.1
ENST00000418627.1 |
TBC1D25
|
TBC1 domain family, member 25 |
| chr16_+_68056844 | 0.14 |
ENST00000565263.1
|
DUS2
|
dihydrouridine synthase 2 |
| chr2_-_65659762 | 0.14 |
ENST00000440972.1
|
SPRED2
|
sprouty-related, EVH1 domain containing 2 |
| chr7_-_93633684 | 0.14 |
ENST00000222547.3
ENST00000425626.1 |
BET1
|
Bet1 golgi vesicular membrane trafficking protein |
| chr9_-_100684845 | 0.13 |
ENST00000375119.3
|
C9orf156
|
chromosome 9 open reading frame 156 |
| chr16_+_56225248 | 0.13 |
ENST00000262493.6
|
GNAO1
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O |
| chr6_-_31940065 | 0.12 |
ENST00000375349.3
ENST00000337523.5 |
DXO
|
decapping exoribonuclease |
| chr1_+_151682909 | 0.12 |
ENST00000326413.3
ENST00000442233.2 |
RIIAD1
AL589765.1
|
regulatory subunit of type II PKA R-subunit (RIIa) domain containing 1 Uncharacterized protein; cDNA FLJ36032 fis, clone TESTI2017069 |
| chr5_-_175964366 | 0.11 |
ENST00000274811.4
|
RNF44
|
ring finger protein 44 |
| chr16_+_68057179 | 0.10 |
ENST00000567100.1
ENST00000432752.1 ENST00000569289.1 ENST00000564781.1 |
DUS2
|
dihydrouridine synthase 2 |
| chr15_-_34394119 | 0.08 |
ENST00000256545.4
|
EMC7
|
ER membrane protein complex subunit 7 |
| chr16_+_68057153 | 0.08 |
ENST00000358896.6
ENST00000568099.2 |
DUS2
|
dihydrouridine synthase 2 |
| chr13_-_36871886 | 0.08 |
ENST00000491049.2
ENST00000503173.1 ENST00000239860.6 ENST00000379862.2 ENST00000239859.7 ENST00000379864.2 ENST00000510088.1 ENST00000554962.1 ENST00000511166.1 |
CCDC169
SOHLH2
CCDC169-SOHLH2
|
coiled-coil domain containing 169 spermatogenesis and oogenesis specific basic helix-loop-helix 2 CCDC169-SOHLH2 readthrough |
| chr3_+_128598433 | 0.07 |
ENST00000308982.7
ENST00000514336.1 |
ACAD9
|
acyl-CoA dehydrogenase family, member 9 |
| chr17_-_7835228 | 0.07 |
ENST00000303731.4
ENST00000571947.1 ENST00000540486.1 ENST00000572656.1 |
TRAPPC1
|
trafficking protein particle complex 1 |
| chr10_-_128975273 | 0.07 |
ENST00000424811.2
|
FAM196A
|
family with sequence similarity 196, member A |
| chr6_-_106773491 | 0.06 |
ENST00000360666.4
|
ATG5
|
autophagy related 5 |
| chr15_-_34394008 | 0.06 |
ENST00000527822.1
ENST00000528949.1 |
EMC7
|
ER membrane protein complex subunit 7 |
| chr11_+_102217936 | 0.06 |
ENST00000532832.1
ENST00000530675.1 ENST00000533742.1 ENST00000227758.2 ENST00000532672.1 ENST00000531259.1 ENST00000527465.1 |
BIRC2
|
baculoviral IAP repeat containing 2 |
| chr6_-_31939734 | 0.06 |
ENST00000375356.3
|
DXO
|
decapping exoribonuclease |
| chr2_-_65357225 | 0.06 |
ENST00000398529.3
ENST00000409751.1 ENST00000356214.7 ENST00000409892.1 ENST00000409784.3 |
RAB1A
|
RAB1A, member RAS oncogene family |
| chr1_+_70671363 | 0.06 |
ENST00000370951.1
ENST00000370950.3 ENST00000405432.1 ENST00000454435.2 |
SRSF11
|
serine/arginine-rich splicing factor 11 |
| chr4_-_39367949 | 0.05 |
ENST00000503784.1
ENST00000349703.2 ENST00000381897.1 |
RFC1
|
replication factor C (activator 1) 1, 145kDa |
| chr11_-_85376121 | 0.05 |
ENST00000527447.1
|
CREBZF
|
CREB/ATF bZIP transcription factor |
| chr6_-_106773610 | 0.04 |
ENST00000369076.3
ENST00000369070.1 |
ATG5
|
autophagy related 5 |
| chr6_-_91006627 | 0.03 |
ENST00000537989.1
|
BACH2
|
BTB and CNC homology 1, basic leucine zipper transcription factor 2 |
| chr12_+_10365404 | 0.03 |
ENST00000266458.5
ENST00000421801.2 ENST00000544284.1 ENST00000545047.1 ENST00000543602.1 ENST00000545887.1 |
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr2_-_202645835 | 0.03 |
ENST00000264276.6
|
ALS2
|
amyotrophic lateral sclerosis 2 (juvenile) |
| chr15_-_66790146 | 0.02 |
ENST00000316634.5
|
SNAPC5
|
small nuclear RNA activating complex, polypeptide 5, 19kDa |
| chr3_+_184056614 | 0.02 |
ENST00000453072.1
|
FAM131A
|
family with sequence similarity 131, member A |
| chr2_-_43823093 | 0.02 |
ENST00000405006.4
|
THADA
|
thyroid adenoma associated |
| chr7_-_93633658 | 0.02 |
ENST00000433727.1
|
BET1
|
Bet1 golgi vesicular membrane trafficking protein |
| chr18_+_76829441 | 0.01 |
ENST00000458297.2
|
ATP9B
|
ATPase, class II, type 9B |
| chr4_+_170581213 | 0.01 |
ENST00000507875.1
|
CLCN3
|
chloride channel, voltage-sensitive 3 |
| chr19_-_18392422 | 0.00 |
ENST00000252818.3
|
JUND
|
jun D proto-oncogene |
Network of associatons between targets according to the STRING database.
First level regulatory network of BATF3
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.1 | 0.4 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.1 | 0.3 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.1 | 0.3 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.1 | 0.9 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.1 | 0.2 | GO:2000987 | hormone-mediated apoptotic signaling pathway(GO:0008628) cellular response to cocaine(GO:0071314) positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.1 | 0.5 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.1 | 0.2 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.0 | 0.1 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.0 | 0.5 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.0 | 0.3 | GO:0002943 | tRNA dihydrouridine synthesis(GO:0002943) |
| 0.0 | 0.2 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.0 | 0.2 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.1 | GO:0060296 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 0.1 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.0 | 0.6 | GO:0007567 | parturition(GO:0007567) |
| 0.0 | 0.1 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.0 | 0.2 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.2 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.0 | 0.2 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.1 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.3 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.1 | GO:0072546 | ER membrane protein complex(GO:0072546) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.1 | 0.2 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.0 | 0.2 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.3 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.4 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.0 | 0.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.1 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.2 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.2 | GO:0030957 | Tat protein binding(GO:0030957) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.4 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |