Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
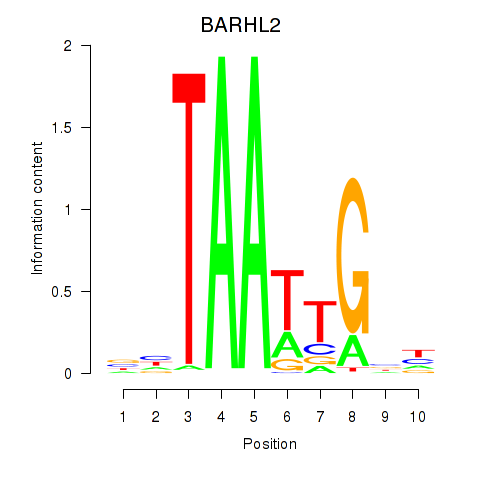
Results for BARHL2
Z-value: 0.76
Transcription factors associated with BARHL2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
BARHL2
|
ENSG00000143032.7 | BarH like homeobox 2 |
Activity profile of BARHL2 motif
Sorted Z-values of BARHL2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_105416039 | 0.52 |
ENST00000394767.2
|
CXXC4
|
CXXC finger protein 4 |
| chr12_-_71551868 | 0.36 |
ENST00000247829.3
|
TSPAN8
|
tetraspanin 8 |
| chr11_-_121986923 | 0.35 |
ENST00000560104.1
|
BLID
|
BH3-like motif containing, cell death inducer |
| chr5_+_81601166 | 0.33 |
ENST00000439350.1
|
ATP6AP1L
|
ATPase, H+ transporting, lysosomal accessory protein 1-like |
| chr12_-_118628315 | 0.32 |
ENST00000540561.1
|
TAOK3
|
TAO kinase 3 |
| chrX_-_49056635 | 0.32 |
ENST00000472598.1
ENST00000538567.1 ENST00000479808.1 ENST00000263233.4 |
SYP
|
synaptophysin |
| chr2_+_145780725 | 0.31 |
ENST00000451478.1
|
TEX41
|
testis expressed 41 (non-protein coding) |
| chr20_-_45530365 | 0.30 |
ENST00000414085.1
|
RP11-323C15.2
|
RP11-323C15.2 |
| chr14_+_61449076 | 0.29 |
ENST00000526105.1
|
SLC38A6
|
solute carrier family 38, member 6 |
| chr12_-_118628350 | 0.29 |
ENST00000537952.1
ENST00000537822.1 |
TAOK3
|
TAO kinase 3 |
| chr5_+_53686658 | 0.29 |
ENST00000512618.1
|
LINC01033
|
long intergenic non-protein coding RNA 1033 |
| chr18_+_20714525 | 0.28 |
ENST00000400473.2
|
CABLES1
|
Cdk5 and Abl enzyme substrate 1 |
| chr20_+_56964169 | 0.28 |
ENST00000475243.1
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr17_+_56769924 | 0.27 |
ENST00000461271.1
ENST00000583539.1 ENST00000337432.4 ENST00000421782.2 |
RAD51C
|
RAD51 paralog C |
| chr1_+_59486059 | 0.26 |
ENST00000447329.1
|
RP4-794H19.4
|
RP4-794H19.4 |
| chr1_-_220263096 | 0.25 |
ENST00000463953.1
ENST00000354807.3 ENST00000414869.2 ENST00000498237.2 ENST00000498791.2 ENST00000544404.1 ENST00000480959.2 ENST00000322067.7 |
BPNT1
|
3'(2'), 5'-bisphosphate nucleotidase 1 |
| chr14_+_32798547 | 0.25 |
ENST00000557354.1
ENST00000557102.1 ENST00000557272.1 |
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr8_-_25281747 | 0.24 |
ENST00000421054.2
|
GNRH1
|
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr14_+_39944025 | 0.24 |
ENST00000554328.1
ENST00000556620.1 ENST00000557197.1 |
RP11-111A21.1
|
RP11-111A21.1 |
| chr12_-_54652060 | 0.23 |
ENST00000552562.1
|
CBX5
|
chromobox homolog 5 |
| chrX_-_46187069 | 0.23 |
ENST00000446884.1
|
RP1-30G7.2
|
RP1-30G7.2 |
| chr12_-_71551652 | 0.23 |
ENST00000546561.1
|
TSPAN8
|
tetraspanin 8 |
| chr5_+_150639360 | 0.23 |
ENST00000523004.1
|
GM2A
|
GM2 ganglioside activator |
| chr16_+_69345243 | 0.22 |
ENST00000254950.11
|
VPS4A
|
vacuolar protein sorting 4 homolog A (S. cerevisiae) |
| chr10_-_101825151 | 0.22 |
ENST00000441382.1
|
CPN1
|
carboxypeptidase N, polypeptide 1 |
| chr12_-_65146717 | 0.22 |
ENST00000545273.1
|
GNS
|
glucosamine (N-acetyl)-6-sulfatase |
| chr2_+_87808725 | 0.22 |
ENST00000413202.1
|
LINC00152
|
long intergenic non-protein coding RNA 152 |
| chr15_+_63188009 | 0.21 |
ENST00000557900.1
|
RP11-1069G10.2
|
RP11-1069G10.2 |
| chr17_+_47448102 | 0.21 |
ENST00000576461.1
|
RP11-81K2.1
|
Uncharacterized protein |
| chr16_+_53412368 | 0.21 |
ENST00000565189.1
|
RP11-44F14.2
|
RP11-44F14.2 |
| chr2_-_188312971 | 0.20 |
ENST00000410068.1
ENST00000447403.1 ENST00000410102.1 |
CALCRL
|
calcitonin receptor-like |
| chr20_-_7921090 | 0.20 |
ENST00000378789.3
|
HAO1
|
hydroxyacid oxidase (glycolate oxidase) 1 |
| chr7_-_6066183 | 0.20 |
ENST00000422786.1
|
EIF2AK1
|
eukaryotic translation initiation factor 2-alpha kinase 1 |
| chr1_+_179263308 | 0.19 |
ENST00000426956.1
|
SOAT1
|
sterol O-acyltransferase 1 |
| chr2_-_224467002 | 0.18 |
ENST00000421386.1
ENST00000433889.1 |
SCG2
|
secretogranin II |
| chr1_-_166845515 | 0.18 |
ENST00000367874.4
|
TADA1
|
transcriptional adaptor 1 |
| chr6_+_26087509 | 0.18 |
ENST00000397022.3
ENST00000353147.5 ENST00000352392.4 ENST00000349999.4 ENST00000317896.7 ENST00000357618.5 ENST00000470149.1 ENST00000336625.8 ENST00000461397.1 ENST00000488199.1 |
HFE
|
hemochromatosis |
| chr21_+_17792672 | 0.18 |
ENST00000602620.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr8_+_94752349 | 0.18 |
ENST00000391680.1
|
RBM12B-AS1
|
RBM12B antisense RNA 1 |
| chr9_-_95244781 | 0.18 |
ENST00000375544.3
ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN
|
asporin |
| chr17_-_40288449 | 0.18 |
ENST00000552162.1
ENST00000550504.1 |
RAB5C
|
RAB5C, member RAS oncogene family |
| chr17_-_39093672 | 0.17 |
ENST00000209718.3
ENST00000436344.3 ENST00000485751.1 |
KRT23
|
keratin 23 (histone deacetylase inducible) |
| chr6_+_13182751 | 0.17 |
ENST00000415087.1
|
PHACTR1
|
phosphatase and actin regulator 1 |
| chrX_-_71351678 | 0.17 |
ENST00000609883.1
ENST00000545866.1 |
RGAG4
|
retrotransposon gag domain containing 4 |
| chr1_-_190446759 | 0.17 |
ENST00000367462.3
|
BRINP3
|
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr12_-_27924209 | 0.17 |
ENST00000381273.3
|
MANSC4
|
MANSC domain containing 4 |
| chr18_+_55888767 | 0.17 |
ENST00000431212.2
ENST00000586268.1 ENST00000587190.1 |
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr19_+_48949087 | 0.16 |
ENST00000598711.1
|
GRWD1
|
glutamate-rich WD repeat containing 1 |
| chr20_+_56964253 | 0.16 |
ENST00000395802.3
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr1_+_44444865 | 0.16 |
ENST00000372324.1
|
B4GALT2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chrX_-_100129320 | 0.16 |
ENST00000372966.3
|
NOX1
|
NADPH oxidase 1 |
| chr1_-_101360205 | 0.16 |
ENST00000450240.1
|
EXTL2
|
exostosin-like glycosyltransferase 2 |
| chr4_+_147096837 | 0.16 |
ENST00000296581.5
ENST00000502781.1 |
LSM6
|
LSM6 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr6_+_28249332 | 0.16 |
ENST00000259883.3
|
PGBD1
|
piggyBac transposable element derived 1 |
| chr6_+_28249299 | 0.16 |
ENST00000405948.2
|
PGBD1
|
piggyBac transposable element derived 1 |
| chr3_-_49066811 | 0.15 |
ENST00000442157.1
ENST00000326739.4 |
IMPDH2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2 |
| chr22_+_51176624 | 0.15 |
ENST00000216139.5
ENST00000529621.1 |
ACR
|
acrosin |
| chr5_-_125930929 | 0.15 |
ENST00000553117.1
ENST00000447989.2 ENST00000409134.3 |
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1 |
| chr2_-_87248975 | 0.15 |
ENST00000409310.2
ENST00000355705.3 |
PLGLB1
|
plasminogen-like B1 |
| chr12_+_7013897 | 0.15 |
ENST00000007969.8
ENST00000323702.5 |
LRRC23
|
leucine rich repeat containing 23 |
| chr3_-_180397256 | 0.15 |
ENST00000442201.2
|
CCDC39
|
coiled-coil domain containing 39 |
| chr10_+_103348031 | 0.15 |
ENST00000370151.4
ENST00000370147.1 ENST00000370148.2 |
DPCD
|
deleted in primary ciliary dyskinesia homolog (mouse) |
| chr17_-_47045949 | 0.15 |
ENST00000357424.2
|
GIP
|
gastric inhibitory polypeptide |
| chr4_-_71532339 | 0.15 |
ENST00000254801.4
|
IGJ
|
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr8_-_109799793 | 0.15 |
ENST00000297459.3
|
TMEM74
|
transmembrane protein 74 |
| chr6_-_26189304 | 0.15 |
ENST00000340756.2
|
HIST1H4D
|
histone cluster 1, H4d |
| chr15_+_59910132 | 0.15 |
ENST00000559200.1
|
GCNT3
|
glucosaminyl (N-acetyl) transferase 3, mucin type |
| chrX_-_68385274 | 0.14 |
ENST00000374584.3
ENST00000590146.1 |
PJA1
|
praja ring finger 1, E3 ubiquitin protein ligase |
| chr20_+_45947246 | 0.14 |
ENST00000599904.1
|
AL031666.2
|
HCG2018772; Uncharacterized protein; cDNA FLJ31609 fis, clone NT2RI2002852 |
| chr22_-_31688431 | 0.14 |
ENST00000402249.3
ENST00000443175.1 ENST00000215912.5 ENST00000441972.1 |
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr6_-_30640761 | 0.14 |
ENST00000415603.1
|
DHX16
|
DEAH (Asp-Glu-Ala-His) box polypeptide 16 |
| chr16_+_21244986 | 0.14 |
ENST00000311620.5
|
ANKS4B
|
ankyrin repeat and sterile alpha motif domain containing 4B |
| chr11_-_73587719 | 0.14 |
ENST00000545127.1
ENST00000537289.1 ENST00000355693.4 |
COA4
|
cytochrome c oxidase assembly factor 4 homolog (S. cerevisiae) |
| chr3_+_42190714 | 0.14 |
ENST00000449246.1
|
TRAK1
|
trafficking protein, kinesin binding 1 |
| chr11_+_72983246 | 0.14 |
ENST00000393590.2
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr4_+_86749045 | 0.14 |
ENST00000514229.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr19_+_48949030 | 0.14 |
ENST00000253237.5
|
GRWD1
|
glutamate-rich WD repeat containing 1 |
| chr12_-_118796910 | 0.14 |
ENST00000541186.1
ENST00000539872.1 |
TAOK3
|
TAO kinase 3 |
| chr11_-_115127611 | 0.14 |
ENST00000545094.1
|
CADM1
|
cell adhesion molecule 1 |
| chr22_-_36013368 | 0.14 |
ENST00000442617.1
ENST00000397326.2 ENST00000397328.1 ENST00000451685.1 |
MB
|
myoglobin |
| chr1_-_201140673 | 0.14 |
ENST00000367333.2
|
TMEM9
|
transmembrane protein 9 |
| chr12_+_110011571 | 0.14 |
ENST00000539696.1
ENST00000228510.3 ENST00000392727.3 |
MVK
|
mevalonate kinase |
| chr2_-_114647327 | 0.14 |
ENST00000602760.1
|
RP11-141B14.1
|
RP11-141B14.1 |
| chr5_-_102898465 | 0.14 |
ENST00000507423.1
ENST00000230792.2 |
NUDT12
|
nudix (nucleoside diphosphate linked moiety X)-type motif 12 |
| chr3_-_126327398 | 0.14 |
ENST00000383572.2
|
TXNRD3NB
|
thioredoxin reductase 3 neighbor |
| chr6_+_170863353 | 0.13 |
ENST00000421512.1
|
TBP
|
TATA box binding protein |
| chr11_-_115158193 | 0.13 |
ENST00000543540.1
|
CADM1
|
cell adhesion molecule 1 |
| chrX_+_149867681 | 0.13 |
ENST00000438018.1
ENST00000436701.1 |
MTMR1
|
myotubularin related protein 1 |
| chr2_+_135596180 | 0.13 |
ENST00000283054.4
ENST00000392928.1 |
ACMSD
|
aminocarboxymuconate semialdehyde decarboxylase |
| chr5_+_142286887 | 0.13 |
ENST00000451259.1
|
ARHGAP26
|
Rho GTPase activating protein 26 |
| chr19_-_10445399 | 0.13 |
ENST00000592945.1
|
ICAM3
|
intercellular adhesion molecule 3 |
| chr6_-_11779014 | 0.13 |
ENST00000229583.5
|
ADTRP
|
androgen-dependent TFPI-regulating protein |
| chr2_-_183387064 | 0.13 |
ENST00000536095.1
ENST00000331935.6 ENST00000358139.2 ENST00000456212.1 |
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr18_+_55816546 | 0.13 |
ENST00000435432.2
ENST00000357895.5 ENST00000586263.1 |
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr10_+_98741041 | 0.13 |
ENST00000286067.2
|
C10orf12
|
chromosome 10 open reading frame 12 |
| chr11_-_71823266 | 0.13 |
ENST00000538919.1
ENST00000539395.1 ENST00000542531.1 |
ANAPC15
|
anaphase promoting complex subunit 15 |
| chrX_+_591524 | 0.13 |
ENST00000554971.1
ENST00000381575.1 |
SHOX
|
short stature homeobox |
| chr12_-_10542617 | 0.13 |
ENST00000240618.6
|
KLRK1
|
killer cell lectin-like receptor subfamily K, member 1 |
| chr5_-_74162605 | 0.13 |
ENST00000389156.4
ENST00000510496.1 ENST00000380515.3 |
FAM169A
|
family with sequence similarity 169, member A |
| chr7_+_99717230 | 0.13 |
ENST00000262932.3
|
CNPY4
|
canopy FGF signaling regulator 4 |
| chr16_-_48281305 | 0.13 |
ENST00000356608.2
|
ABCC11
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 11 |
| chr9_+_125486269 | 0.12 |
ENST00000259466.1
|
OR1L4
|
olfactory receptor, family 1, subfamily L, member 4 |
| chr6_+_26199737 | 0.12 |
ENST00000359985.1
|
HIST1H2BF
|
histone cluster 1, H2bf |
| chr11_+_35222629 | 0.12 |
ENST00000526553.1
|
CD44
|
CD44 molecule (Indian blood group) |
| chr12_+_7014064 | 0.12 |
ENST00000443597.2
|
LRRC23
|
leucine rich repeat containing 23 |
| chr19_-_44259136 | 0.12 |
ENST00000270066.6
|
SMG9
|
SMG9 nonsense mediated mRNA decay factor |
| chr22_-_31688381 | 0.12 |
ENST00000487265.2
|
PIK3IP1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr4_+_71019903 | 0.12 |
ENST00000344526.5
|
C4orf40
|
chromosome 4 open reading frame 40 |
| chr14_-_70883708 | 0.12 |
ENST00000256366.4
|
SYNJ2BP
|
synaptojanin 2 binding protein |
| chrX_-_15288154 | 0.12 |
ENST00000380483.3
ENST00000380485.3 ENST00000380488.4 |
ASB9
|
ankyrin repeat and SOCS box containing 9 |
| chr3_+_184032419 | 0.12 |
ENST00000352767.3
ENST00000427141.2 |
EIF4G1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr1_-_95391315 | 0.12 |
ENST00000545882.1
ENST00000415017.1 |
CNN3
|
calponin 3, acidic |
| chr1_+_62439037 | 0.12 |
ENST00000545929.1
|
INADL
|
InaD-like (Drosophila) |
| chr18_+_72167096 | 0.12 |
ENST00000324301.8
|
CNDP2
|
CNDP dipeptidase 2 (metallopeptidase M20 family) |
| chr12_+_41136144 | 0.12 |
ENST00000548005.1
ENST00000552248.1 |
CNTN1
|
contactin 1 |
| chr12_-_91505608 | 0.12 |
ENST00000266718.4
|
LUM
|
lumican |
| chr12_+_122326662 | 0.12 |
ENST00000261817.2
ENST00000538613.1 ENST00000542602.1 |
PSMD9
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 9 |
| chr15_+_40674920 | 0.12 |
ENST00000416151.2
ENST00000249776.8 |
KNSTRN
|
kinetochore-localized astrin/SPAG5 binding protein |
| chr3_+_173116225 | 0.11 |
ENST00000457714.1
|
NLGN1
|
neuroligin 1 |
| chr17_-_17184605 | 0.11 |
ENST00000268717.5
|
COPS3
|
COP9 signalosome subunit 3 |
| chr3_-_87842631 | 0.11 |
ENST00000462792.1
|
RP11-451B8.1
|
RP11-451B8.1 |
| chr2_+_145780767 | 0.11 |
ENST00000599358.1
ENST00000596278.1 ENST00000596747.1 ENST00000608652.1 ENST00000609705.1 ENST00000608432.1 ENST00000596970.1 ENST00000602041.1 ENST00000601578.1 ENST00000596034.1 ENST00000414195.2 ENST00000594837.1 |
TEX41
|
testis expressed 41 (non-protein coding) |
| chr12_-_10978957 | 0.11 |
ENST00000240619.2
|
TAS2R10
|
taste receptor, type 2, member 10 |
| chr5_-_139726181 | 0.11 |
ENST00000507104.1
ENST00000230990.6 |
HBEGF
|
heparin-binding EGF-like growth factor |
| chr12_-_49245916 | 0.11 |
ENST00000552512.1
ENST00000551468.1 |
DDX23
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 23 |
| chr19_-_6057282 | 0.11 |
ENST00000592281.1
|
RFX2
|
regulatory factor X, 2 (influences HLA class II expression) |
| chr7_-_112635675 | 0.11 |
ENST00000447785.1
ENST00000451962.1 |
AC018464.3
|
AC018464.3 |
| chr14_+_32798462 | 0.11 |
ENST00000280979.4
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr11_-_17229480 | 0.11 |
ENST00000532035.1
ENST00000540361.1 |
PIK3C2A
|
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 alpha |
| chr4_-_142253761 | 0.11 |
ENST00000511213.1
|
RP11-362F19.1
|
RP11-362F19.1 |
| chr2_+_149402989 | 0.11 |
ENST00000397424.2
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr1_-_109399682 | 0.11 |
ENST00000369995.3
ENST00000370001.3 |
AKNAD1
|
AKNA domain containing 1 |
| chr16_-_46655538 | 0.11 |
ENST00000303383.3
|
SHCBP1
|
SHC SH2-domain binding protein 1 |
| chr12_+_58087738 | 0.11 |
ENST00000552285.1
|
OS9
|
osteosarcoma amplified 9, endoplasmic reticulum lectin |
| chr17_-_18430160 | 0.11 |
ENST00000392176.3
|
FAM106A
|
family with sequence similarity 106, member A |
| chr5_+_136070614 | 0.11 |
ENST00000502421.1
|
CTB-1I21.1
|
CTB-1I21.1 |
| chr5_+_140855495 | 0.11 |
ENST00000308177.3
|
PCDHGC3
|
protocadherin gamma subfamily C, 3 |
| chr17_-_48785216 | 0.11 |
ENST00000285243.6
|
ANKRD40
|
ankyrin repeat domain 40 |
| chr17_-_39203519 | 0.11 |
ENST00000542137.1
ENST00000391419.3 |
KRTAP2-1
|
keratin associated protein 2-1 |
| chr15_+_40675132 | 0.11 |
ENST00000608100.1
ENST00000557920.1 |
KNSTRN
|
kinetochore-localized astrin/SPAG5 binding protein |
| chr7_+_148982396 | 0.11 |
ENST00000418158.2
|
ZNF783
|
zinc finger family member 783 |
| chr14_-_78083112 | 0.11 |
ENST00000216484.2
|
SPTLC2
|
serine palmitoyltransferase, long chain base subunit 2 |
| chr7_-_130080977 | 0.11 |
ENST00000223208.5
|
CEP41
|
centrosomal protein 41kDa |
| chr11_+_34664014 | 0.10 |
ENST00000527935.1
|
EHF
|
ets homologous factor |
| chr22_-_40929812 | 0.10 |
ENST00000422851.1
|
MKL1
|
megakaryoblastic leukemia (translocation) 1 |
| chr14_+_61449197 | 0.10 |
ENST00000533744.2
|
SLC38A6
|
solute carrier family 38, member 6 |
| chr11_+_93479588 | 0.10 |
ENST00000526335.1
|
C11orf54
|
chromosome 11 open reading frame 54 |
| chr12_+_54402790 | 0.10 |
ENST00000040584.4
|
HOXC8
|
homeobox C8 |
| chr20_-_25371412 | 0.10 |
ENST00000339157.5
ENST00000376542.3 |
ABHD12
|
abhydrolase domain containing 12 |
| chr13_-_41706864 | 0.10 |
ENST00000379485.1
ENST00000499385.2 |
KBTBD6
|
kelch repeat and BTB (POZ) domain containing 6 |
| chr4_-_70518941 | 0.10 |
ENST00000286604.4
ENST00000505512.1 ENST00000514019.1 |
UGT2A1
UGT2A1
|
UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus |
| chr4_-_44653636 | 0.10 |
ENST00000415895.4
ENST00000332990.5 |
YIPF7
|
Yip1 domain family, member 7 |
| chr1_+_207277590 | 0.10 |
ENST00000367070.3
|
C4BPA
|
complement component 4 binding protein, alpha |
| chr12_-_27235447 | 0.10 |
ENST00000429849.2
|
C12orf71
|
chromosome 12 open reading frame 71 |
| chr15_+_76016293 | 0.10 |
ENST00000332145.2
|
ODF3L1
|
outer dense fiber of sperm tails 3-like 1 |
| chr2_+_152214098 | 0.10 |
ENST00000243347.3
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr3_-_178103144 | 0.10 |
ENST00000417383.1
ENST00000418585.1 ENST00000411727.1 ENST00000439810.1 |
RP11-33A14.1
|
RP11-33A14.1 |
| chr2_-_55496174 | 0.10 |
ENST00000417363.1
ENST00000412530.1 ENST00000394600.3 ENST00000366137.2 ENST00000420637.1 |
MTIF2
|
mitochondrial translational initiation factor 2 |
| chr6_-_76203454 | 0.10 |
ENST00000237172.7
|
FILIP1
|
filamin A interacting protein 1 |
| chr1_+_110993795 | 0.10 |
ENST00000271331.3
|
PROK1
|
prokineticin 1 |
| chr9_+_125027127 | 0.10 |
ENST00000441707.1
ENST00000373723.5 ENST00000373729.1 |
MRRF
|
mitochondrial ribosome recycling factor |
| chr4_+_56814968 | 0.10 |
ENST00000422247.2
|
CEP135
|
centrosomal protein 135kDa |
| chr2_+_158114051 | 0.10 |
ENST00000259056.4
|
GALNT5
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) |
| chr19_+_36606354 | 0.10 |
ENST00000589996.1
ENST00000591296.1 |
TBCB
|
tubulin folding cofactor B |
| chr6_+_34204642 | 0.10 |
ENST00000347617.6
ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1
|
high mobility group AT-hook 1 |
| chr6_+_43603552 | 0.10 |
ENST00000372171.4
|
MAD2L1BP
|
MAD2L1 binding protein |
| chr21_+_43619796 | 0.10 |
ENST00000398457.2
|
ABCG1
|
ATP-binding cassette, sub-family G (WHITE), member 1 |
| chr5_+_60933634 | 0.10 |
ENST00000505642.1
|
C5orf64
|
chromosome 5 open reading frame 64 |
| chr2_-_203735586 | 0.10 |
ENST00000454326.1
ENST00000432273.1 ENST00000450143.1 ENST00000411681.1 |
ICA1L
|
islet cell autoantigen 1,69kDa-like |
| chr3_-_25915189 | 0.10 |
ENST00000451284.1
|
LINC00692
|
long intergenic non-protein coding RNA 692 |
| chr10_+_94594351 | 0.10 |
ENST00000371552.4
|
EXOC6
|
exocyst complex component 6 |
| chr12_+_104697504 | 0.10 |
ENST00000527879.1
|
EID3
|
EP300 interacting inhibitor of differentiation 3 |
| chr2_-_220110111 | 0.10 |
ENST00000428427.1
ENST00000356283.3 ENST00000432839.1 ENST00000424620.1 |
GLB1L
|
galactosidase, beta 1-like |
| chrX_+_70521584 | 0.09 |
ENST00000373829.3
ENST00000538820.1 |
ITGB1BP2
|
integrin beta 1 binding protein (melusin) 2 |
| chr2_+_196313239 | 0.09 |
ENST00000413290.1
|
AC064834.1
|
AC064834.1 |
| chr1_-_232651312 | 0.09 |
ENST00000262861.4
|
SIPA1L2
|
signal-induced proliferation-associated 1 like 2 |
| chr16_-_69418553 | 0.09 |
ENST00000569542.2
|
TERF2
|
telomeric repeat binding factor 2 |
| chr17_-_39211463 | 0.09 |
ENST00000542910.1
ENST00000398477.1 |
KRTAP2-2
|
keratin associated protein 2-2 |
| chr11_+_77300669 | 0.09 |
ENST00000313578.3
|
AQP11
|
aquaporin 11 |
| chr3_+_193965426 | 0.09 |
ENST00000455821.1
|
RP11-513G11.3
|
RP11-513G11.3 |
| chr19_-_41870026 | 0.09 |
ENST00000243578.3
|
B9D2
|
B9 protein domain 2 |
| chr9_+_116263778 | 0.09 |
ENST00000394646.3
|
RGS3
|
regulator of G-protein signaling 3 |
| chr11_+_110001723 | 0.09 |
ENST00000528673.1
|
ZC3H12C
|
zinc finger CCCH-type containing 12C |
| chr17_+_37856299 | 0.09 |
ENST00000269571.5
|
ERBB2
|
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 |
| chr12_+_58087901 | 0.09 |
ENST00000315970.7
ENST00000547079.1 ENST00000439210.2 ENST00000389146.6 ENST00000413095.2 ENST00000551035.1 ENST00000257966.8 ENST00000435406.2 ENST00000550372.1 ENST00000389142.5 |
OS9
|
osteosarcoma amplified 9, endoplasmic reticulum lectin |
| chrX_-_74145273 | 0.09 |
ENST00000055682.6
|
KIAA2022
|
KIAA2022 |
| chr7_+_128784712 | 0.09 |
ENST00000289407.4
|
TSPAN33
|
tetraspanin 33 |
| chr4_+_169013666 | 0.09 |
ENST00000359299.3
|
ANXA10
|
annexin A10 |
| chr3_+_38017264 | 0.09 |
ENST00000436654.1
|
CTDSPL
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
| chr15_+_89164520 | 0.09 |
ENST00000332810.3
|
AEN
|
apoptosis enhancing nuclease |
| chr20_+_55043647 | 0.09 |
ENST00000023939.4
ENST00000395881.3 ENST00000357348.5 ENST00000449062.1 ENST00000435342.2 |
RTFDC1
|
replication termination factor 2 domain containing 1 |
| chr5_+_179159813 | 0.09 |
ENST00000292599.3
|
MAML1
|
mastermind-like 1 (Drosophila) |
| chr4_+_169418195 | 0.09 |
ENST00000261509.6
ENST00000335742.7 |
PALLD
|
palladin, cytoskeletal associated protein |
| chr13_-_30160925 | 0.09 |
ENST00000450494.1
|
SLC7A1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr1_+_92545862 | 0.09 |
ENST00000370382.3
ENST00000342818.3 |
BTBD8
|
BTB (POZ) domain containing 8 |
| chr1_+_32538492 | 0.09 |
ENST00000336294.5
|
TMEM39B
|
transmembrane protein 39B |
| chr1_-_93257951 | 0.09 |
ENST00000543509.1
ENST00000370331.1 ENST00000540033.1 |
EVI5
|
ecotropic viral integration site 5 |
| chr5_+_140602904 | 0.09 |
ENST00000515856.2
ENST00000239449.4 |
PCDHB14
|
protocadherin beta 14 |
| chr6_-_75960024 | 0.09 |
ENST00000370081.2
|
COX7A2
|
cytochrome c oxidase subunit VIIa polypeptide 2 (liver) |
| chr8_+_107738240 | 0.09 |
ENST00000449762.2
ENST00000297447.6 |
OXR1
|
oxidation resistance 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of BARHL2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | GO:0090402 | senescence-associated heterochromatin focus assembly(GO:0035986) oncogene-induced cell senescence(GO:0090402) |
| 0.1 | 0.2 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.1 | 0.2 | GO:0030070 | insulin processing(GO:0030070) |
| 0.1 | 0.5 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.1 | 0.2 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.1 | 0.4 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.1 | 0.2 | GO:0002590 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 0.1 | 0.2 | GO:0060380 | regulation of single-stranded telomeric DNA binding(GO:0060380) positive regulation of single-stranded telomeric DNA binding(GO:0060381) |
| 0.1 | 0.3 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.1 | 0.3 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 | 0.2 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.0 | 0.3 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.2 | GO:0000354 | cis assembly of pre-catalytic spliceosome(GO:0000354) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.1 | GO:0006742 | NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.0 | 0.2 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.0 | 0.4 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.1 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.0 | 0.2 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) |
| 0.0 | 0.1 | GO:0099543 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.0 | 0.2 | GO:0019285 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.0 | 0.3 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 0.0 | 0.1 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.0 | 0.1 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.1 | GO:1904106 | protein localization to microvillus(GO:1904106) |
| 0.0 | 0.1 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 0.2 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.0 | 0.0 | GO:0061145 | bronchus cartilage development(GO:0060532) lung smooth muscle development(GO:0061145) |
| 0.0 | 0.1 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.3 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.1 | GO:0002528 | regulation of vascular permeability involved in acute inflammatory response(GO:0002528) |
| 0.0 | 0.2 | GO:0009441 | glycolate metabolic process(GO:0009441) |
| 0.0 | 0.1 | GO:0032207 | regulation of telomere maintenance via recombination(GO:0032207) negative regulation of telomere maintenance via recombination(GO:0032208) negative regulation of single strand break repair(GO:1903517) negative regulation of beta-galactosidase activity(GO:1903770) telomere single strand break repair(GO:1903823) negative regulation of telomere single strand break repair(GO:1903824) |
| 0.0 | 0.2 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.1 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.4 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.1 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.2 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.2 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 0.1 | GO:0060922 | cardiac septum cell differentiation(GO:0003292) atrioventricular node cell differentiation(GO:0060922) atrioventricular node cell development(GO:0060928) |
| 0.0 | 0.2 | GO:0034627 | 'de novo' NAD biosynthetic process(GO:0034627) |
| 0.0 | 0.2 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.8 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.1 | GO:1903045 | neural crest cell migration involved in sympathetic nervous system development(GO:1903045) |
| 0.0 | 0.1 | GO:0006173 | dADP biosynthetic process(GO:0006173) |
| 0.0 | 0.2 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.0 | 0.1 | GO:0070124 | mitochondrial translational initiation(GO:0070124) |
| 0.0 | 0.1 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.0 | 0.1 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.1 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.2 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.2 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.1 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.0 | 0.1 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 0.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.1 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.2 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.1 | GO:0044026 | DNA hypermethylation(GO:0044026) |
| 0.0 | 0.3 | GO:0015865 | purine nucleotide transport(GO:0015865) |
| 0.0 | 0.1 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.1 | GO:0021678 | third ventricle development(GO:0021678) |
| 0.0 | 0.0 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) positive regulation of histone H3-K79 methylation(GO:2001162) |
| 0.0 | 0.0 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) gall bladder development(GO:0061010) |
| 0.0 | 0.0 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.0 | 0.0 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.3 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.1 | GO:1903826 | arginine transmembrane transport(GO:1903826) |
| 0.0 | 0.1 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.1 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.0 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) |
| 0.0 | 0.1 | GO:1900827 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.1 | GO:0071503 | response to heparin(GO:0071503) |
| 0.0 | 0.1 | GO:0015692 | lead ion transport(GO:0015692) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.0 | 0.3 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.1 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 0.4 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.0 | GO:0036019 | endolysosome(GO:0036019) endolysosome lumen(GO:0036021) |
| 0.0 | 0.2 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.1 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.2 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.2 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.3 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.1 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.1 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.3 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.0 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 0.1 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.0 | 0.1 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 0.2 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.2 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.1 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.0 | 0.4 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.3 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.2 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.1 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.1 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.1 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0070826 | paraferritin complex(GO:0070826) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 0.3 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.1 | 0.3 | GO:0008441 | 3'(2'),5'-bisphosphate nucleotidase activity(GO:0008441) |
| 0.1 | 0.2 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.1 | 0.2 | GO:0034736 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.1 | 0.2 | GO:0035248 | alpha-1,4-N-acetylgalactosaminyltransferase activity(GO:0035248) |
| 0.1 | 0.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.1 | GO:0047225 | acetylgalactosaminyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0047225) |
| 0.0 | 0.2 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.2 | GO:0004040 | amidase activity(GO:0004040) |
| 0.0 | 0.2 | GO:0001034 | RNA polymerase III transcription factor activity, sequence-specific DNA binding(GO:0001034) |
| 0.0 | 0.2 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.2 | GO:0097642 | calcitonin family receptor activity(GO:0097642) |
| 0.0 | 0.2 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.1 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) |
| 0.0 | 0.2 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.2 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.1 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.1 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.2 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.3 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.4 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.3 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.1 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.0 | 0.1 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.0 | 0.1 | GO:0047298 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.1 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.0 | 0.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.3 | GO:0015216 | purine nucleotide transmembrane transporter activity(GO:0015216) |
| 0.0 | 0.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0045029 | uridine nucleotide receptor activity(GO:0015065) UDP-activated nucleotide receptor activity(GO:0045029) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.1 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.0 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.0 | 0.1 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 0.2 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.3 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.1 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0015094 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.2 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.1 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.5 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |