Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
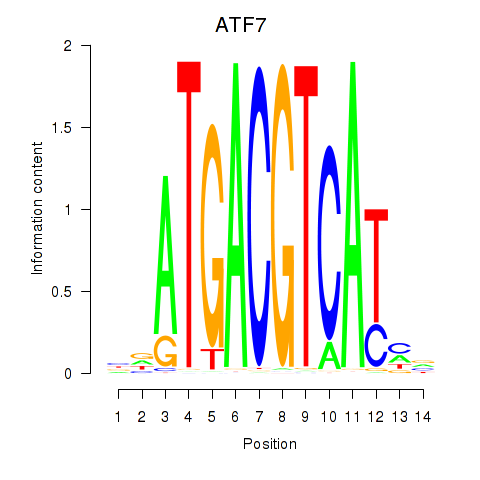
Results for ATF7
Z-value: 0.65
Transcription factors associated with ATF7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ATF7
|
ENSG00000170653.14 | activating transcription factor 7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ATF7 | hg19_v2_chr12_-_54020107_54020199 | 0.94 | 5.7e-02 | Click! |
Activity profile of ATF7 motif
Sorted Z-values of ATF7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_153750622 | 0.45 |
ENST00000532853.1
|
SLC27A3
|
solute carrier family 27 (fatty acid transporter), member 3 |
| chr19_+_47616682 | 0.41 |
ENST00000594526.1
|
SAE1
|
SUMO1 activating enzyme subunit 1 |
| chr12_+_10365082 | 0.39 |
ENST00000545859.1
|
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr22_+_37959647 | 0.39 |
ENST00000415670.1
|
CDC42EP1
|
CDC42 effector protein (Rho GTPase binding) 1 |
| chr2_+_74781828 | 0.37 |
ENST00000340004.6
|
DOK1
|
docking protein 1, 62kDa (downstream of tyrosine kinase 1) |
| chr17_-_20946710 | 0.35 |
ENST00000584538.1
|
USP22
|
ubiquitin specific peptidase 22 |
| chr7_-_45026159 | 0.34 |
ENST00000584327.1
ENST00000438705.3 |
SNHG15
|
small nucleolar RNA host gene 15 (non-protein coding) |
| chr11_-_9482010 | 0.34 |
ENST00000596206.1
|
AC132192.1
|
LOC644656 protein; Uncharacterized protein |
| chr9_-_77643307 | 0.33 |
ENST00000376834.3
ENST00000376830.3 |
C9orf41
|
chromosome 9 open reading frame 41 |
| chr6_-_26285737 | 0.32 |
ENST00000377727.1
ENST00000289352.1 |
HIST1H4H
|
histone cluster 1, H4h |
| chr7_-_45026200 | 0.31 |
ENST00000577700.1
ENST00000580458.1 ENST00000579383.1 ENST00000584686.1 ENST00000585030.1 ENST00000582727.1 |
SNHG15
|
small nucleolar RNA host gene 15 (non-protein coding) |
| chr11_-_28129656 | 0.31 |
ENST00000263181.6
|
KIF18A
|
kinesin family member 18A |
| chr14_+_105219437 | 0.29 |
ENST00000329967.6
ENST00000347067.5 ENST00000553810.1 |
SIVA1
|
SIVA1, apoptosis-inducing factor |
| chr4_-_104119528 | 0.29 |
ENST00000380026.3
ENST00000503705.1 ENST00000265148.3 |
CENPE
|
centromere protein E, 312kDa |
| chr10_+_134351319 | 0.28 |
ENST00000368594.3
ENST00000368593.3 |
INPP5A
|
inositol polyphosphate-5-phosphatase, 40kDa |
| chr15_-_91537723 | 0.27 |
ENST00000394249.3
ENST00000559811.1 ENST00000442656.2 ENST00000557905.1 ENST00000361919.3 |
PRC1
|
protein regulator of cytokinesis 1 |
| chr4_-_122744998 | 0.26 |
ENST00000274026.5
|
CCNA2
|
cyclin A2 |
| chr7_+_100464760 | 0.25 |
ENST00000200457.4
|
TRIP6
|
thyroid hormone receptor interactor 6 |
| chr17_-_43209862 | 0.25 |
ENST00000322765.5
|
PLCD3
|
phospholipase C, delta 3 |
| chr6_-_34855773 | 0.25 |
ENST00000420584.2
ENST00000361288.4 |
TAF11
|
TAF11 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 28kDa |
| chr3_-_134092561 | 0.25 |
ENST00000510560.1
ENST00000504234.1 ENST00000515172.1 |
AMOTL2
|
angiomotin like 2 |
| chr8_+_98788003 | 0.25 |
ENST00000521545.2
|
LAPTM4B
|
lysosomal protein transmembrane 4 beta |
| chr7_-_124569991 | 0.24 |
ENST00000446993.1
ENST00000357628.3 ENST00000393329.1 |
POT1
|
protection of telomeres 1 |
| chr7_+_140396946 | 0.24 |
ENST00000476470.1
ENST00000471136.1 |
NDUFB2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2, 8kDa |
| chr19_+_59055412 | 0.24 |
ENST00000593582.1
|
TRIM28
|
tripartite motif containing 28 |
| chr4_+_1873155 | 0.24 |
ENST00000507820.1
ENST00000514045.1 |
WHSC1
|
Wolf-Hirschhorn syndrome candidate 1 |
| chr4_+_85504075 | 0.23 |
ENST00000295887.5
|
CDS1
|
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1 |
| chr3_+_99536663 | 0.23 |
ENST00000421999.2
ENST00000463526.1 |
CMSS1
|
cms1 ribosomal small subunit homolog (yeast) |
| chr3_+_44803209 | 0.23 |
ENST00000326047.4
|
KIF15
|
kinesin family member 15 |
| chr14_+_68086515 | 0.23 |
ENST00000261783.3
|
ARG2
|
arginase 2 |
| chr1_+_110527308 | 0.23 |
ENST00000369799.5
|
AHCYL1
|
adenosylhomocysteinase-like 1 |
| chr7_+_102292796 | 0.23 |
ENST00000436228.2
|
SPDYE2B
|
speedy/RINGO cell cycle regulator family member E2B |
| chr6_+_28048753 | 0.22 |
ENST00000377325.1
|
ZNF165
|
zinc finger protein 165 |
| chr9_-_34665983 | 0.22 |
ENST00000416454.1
ENST00000544078.2 ENST00000421828.2 ENST00000423809.1 |
RP11-195F19.5
|
HCG2040265, isoform CRA_a; Uncharacterized protein; cDNA FLJ50015 |
| chr6_+_30749649 | 0.21 |
ENST00000422944.1
|
HCG20
|
HLA complex group 20 (non-protein coding) |
| chr14_+_61447832 | 0.21 |
ENST00000354886.2
ENST00000267488.4 |
SLC38A6
|
solute carrier family 38, member 6 |
| chr19_-_39402798 | 0.21 |
ENST00000571838.1
|
CTC-360G5.1
|
coiled-coil glutamate-rich protein 2 |
| chr4_+_113558272 | 0.21 |
ENST00000509061.1
ENST00000508577.1 ENST00000513553.1 |
LARP7
|
La ribonucleoprotein domain family, member 7 |
| chr1_+_85527987 | 0.21 |
ENST00000326813.8
ENST00000294664.6 ENST00000528899.1 |
WDR63
|
WD repeat domain 63 |
| chr8_-_95274536 | 0.20 |
ENST00000297596.2
ENST00000396194.2 |
GEM
|
GTP binding protein overexpressed in skeletal muscle |
| chr10_+_99496872 | 0.20 |
ENST00000337540.7
ENST00000357540.4 ENST00000370613.3 ENST00000370610.3 ENST00000393677.4 ENST00000453958.2 ENST00000359980.3 |
ZFYVE27
|
zinc finger, FYVE domain containing 27 |
| chr2_-_98280383 | 0.20 |
ENST00000289228.5
|
ACTR1B
|
ARP1 actin-related protein 1 homolog B, centractin beta (yeast) |
| chr20_+_42295745 | 0.20 |
ENST00000396863.4
ENST00000217026.4 |
MYBL2
|
v-myb avian myeloblastosis viral oncogene homolog-like 2 |
| chr5_-_138739739 | 0.20 |
ENST00000514983.1
ENST00000507779.2 ENST00000451821.2 ENST00000450845.2 ENST00000509959.1 ENST00000302091.5 |
SPATA24
|
spermatogenesis associated 24 |
| chr20_+_5892147 | 0.19 |
ENST00000455042.1
|
CHGB
|
chromogranin B (secretogranin 1) |
| chr1_-_245026388 | 0.19 |
ENST00000440865.1
|
HNRNPU
|
heterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A) |
| chr12_-_52604607 | 0.19 |
ENST00000551894.1
ENST00000553017.1 |
C12orf80
|
chromosome 12 open reading frame 80 |
| chr6_+_126661253 | 0.19 |
ENST00000368326.1
ENST00000368325.1 ENST00000368328.4 |
CENPW
|
centromere protein W |
| chr3_-_154042235 | 0.19 |
ENST00000308361.6
ENST00000496811.1 ENST00000544526.1 |
DHX36
|
DEAH (Asp-Glu-Ala-His) box polypeptide 36 |
| chr14_-_102552659 | 0.19 |
ENST00000441629.2
|
HSP90AA1
|
heat shock protein 90kDa alpha (cytosolic), class A member 1 |
| chr6_+_150690133 | 0.18 |
ENST00000392255.3
ENST00000500320.3 |
IYD
|
iodotyrosine deiodinase |
| chr5_+_138210919 | 0.18 |
ENST00000522013.1
ENST00000520260.1 ENST00000523298.1 ENST00000520865.1 ENST00000519634.1 ENST00000517533.1 ENST00000523685.1 ENST00000519768.1 ENST00000517656.1 ENST00000521683.1 ENST00000521640.1 ENST00000519116.1 |
CTNNA1
|
catenin (cadherin-associated protein), alpha 1, 102kDa |
| chr3_+_51851612 | 0.18 |
ENST00000456080.1
|
IQCF3
|
IQ motif containing F3 |
| chr6_+_139456226 | 0.18 |
ENST00000367658.2
|
HECA
|
headcase homolog (Drosophila) |
| chr16_+_19079215 | 0.18 |
ENST00000544894.2
ENST00000561858.1 |
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast) |
| chr11_+_4116005 | 0.18 |
ENST00000300738.5
|
RRM1
|
ribonucleotide reductase M1 |
| chr6_+_30029008 | 0.18 |
ENST00000332435.5
ENST00000376782.2 ENST00000359374.4 ENST00000376785.2 |
ZNRD1
|
zinc ribbon domain containing 1 |
| chr10_-_38265517 | 0.17 |
ENST00000302609.7
|
ZNF25
|
zinc finger protein 25 |
| chrX_-_47479246 | 0.17 |
ENST00000295987.7
ENST00000340666.4 |
SYN1
|
synapsin I |
| chr5_+_131892603 | 0.17 |
ENST00000378823.3
ENST00000265335.6 |
RAD50
|
RAD50 homolog (S. cerevisiae) |
| chr1_+_17531614 | 0.17 |
ENST00000375471.4
|
PADI1
|
peptidyl arginine deiminase, type I |
| chr9_-_111696224 | 0.17 |
ENST00000537196.1
|
IKBKAP
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase complex-associated protein |
| chrX_-_152343394 | 0.17 |
ENST00000370261.1
|
PNMA6B
|
paraneoplastic Ma antigen family member 6B (pseudogene) |
| chr11_-_77185094 | 0.16 |
ENST00000278568.4
ENST00000356341.3 |
PAK1
|
p21 protein (Cdc42/Rac)-activated kinase 1 |
| chr9_+_125027127 | 0.16 |
ENST00000441707.1
ENST00000373723.5 ENST00000373729.1 |
MRRF
|
mitochondrial ribosome recycling factor |
| chr11_+_4116054 | 0.16 |
ENST00000423050.2
|
RRM1
|
ribonucleotide reductase M1 |
| chr17_-_20946338 | 0.16 |
ENST00000261497.4
|
USP22
|
ubiquitin specific peptidase 22 |
| chr11_-_63439013 | 0.16 |
ENST00000398868.3
|
ATL3
|
atlastin GTPase 3 |
| chr14_+_88851874 | 0.16 |
ENST00000393545.4
ENST00000356583.5 ENST00000555401.1 ENST00000553885.1 |
SPATA7
|
spermatogenesis associated 7 |
| chr12_+_56552128 | 0.16 |
ENST00000548580.1
ENST00000293422.5 ENST00000348108.4 ENST00000549017.1 ENST00000549566.1 ENST00000536128.1 ENST00000547649.1 ENST00000547408.1 ENST00000551589.1 ENST00000549392.1 ENST00000548400.1 ENST00000548293.1 |
MYL6
|
myosin, light chain 6, alkali, smooth muscle and non-muscle |
| chr5_+_139055055 | 0.16 |
ENST00000511457.1
|
CXXC5
|
CXXC finger protein 5 |
| chr3_+_10068095 | 0.16 |
ENST00000287647.3
ENST00000383807.1 ENST00000383806.1 ENST00000419585.1 |
FANCD2
|
Fanconi anemia, complementation group D2 |
| chr18_+_72163536 | 0.16 |
ENST00000579847.1
ENST00000583203.1 ENST00000581513.1 ENST00000577600.1 ENST00000579583.1 ENST00000584613.1 |
CNDP2
|
CNDP dipeptidase 2 (metallopeptidase M20 family) |
| chr7_+_30174574 | 0.16 |
ENST00000409688.1
|
C7orf41
|
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr8_-_10697281 | 0.15 |
ENST00000524114.1
ENST00000553390.1 ENST00000554914.1 |
PINX1
SOX7
SOX7
|
PIN2/TERF1 interacting, telomerase inhibitor 1 SRY (sex determining region Y)-box 7 Transcription factor SOX-7; Uncharacterized protein; cDNA FLJ58508, highly similar to Transcription factor SOX-7 |
| chr7_+_140396756 | 0.15 |
ENST00000460088.1
ENST00000472695.1 |
NDUFB2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2, 8kDa |
| chr17_+_68071389 | 0.15 |
ENST00000283936.1
ENST00000392671.1 |
KCNJ16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr22_-_41252962 | 0.15 |
ENST00000216218.3
|
ST13
|
suppression of tumorigenicity 13 (colon carcinoma) (Hsp70 interacting protein) |
| chr5_-_172198190 | 0.15 |
ENST00000239223.3
|
DUSP1
|
dual specificity phosphatase 1 |
| chrX_+_152240819 | 0.15 |
ENST00000421798.3
ENST00000535416.1 |
PNMA6C
PNMA6A
|
paraneoplastic Ma antigen family member 6C paraneoplastic Ma antigen family member 6A |
| chrX_+_155110956 | 0.15 |
ENST00000286448.6
ENST00000262640.6 ENST00000460621.1 |
VAMP7
|
vesicle-associated membrane protein 7 |
| chr4_+_108911036 | 0.15 |
ENST00000505878.1
|
HADH
|
hydroxyacyl-CoA dehydrogenase |
| chr11_+_18344106 | 0.14 |
ENST00000534641.1
ENST00000525831.1 ENST00000265963.4 |
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa |
| chr11_+_28129795 | 0.14 |
ENST00000406787.3
ENST00000342303.5 ENST00000403099.1 ENST00000407364.3 |
METTL15
|
methyltransferase like 15 |
| chr1_-_201140673 | 0.14 |
ENST00000367333.2
|
TMEM9
|
transmembrane protein 9 |
| chr2_+_37571845 | 0.14 |
ENST00000537448.1
|
QPCT
|
glutaminyl-peptide cyclotransferase |
| chr22_+_25003568 | 0.14 |
ENST00000447416.1
|
GGT1
|
gamma-glutamyltransferase 1 |
| chr22_+_32340447 | 0.14 |
ENST00000248975.5
|
YWHAH
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta |
| chr17_+_43209967 | 0.14 |
ENST00000431281.1
ENST00000591859.1 |
ACBD4
|
acyl-CoA binding domain containing 4 |
| chr1_-_70671216 | 0.14 |
ENST00000370952.3
|
LRRC40
|
leucine rich repeat containing 40 |
| chr16_+_81040794 | 0.14 |
ENST00000439957.3
ENST00000393335.3 ENST00000428963.2 ENST00000564669.1 |
CENPN
|
centromere protein N |
| chr14_+_61447927 | 0.13 |
ENST00000451406.1
|
SLC38A6
|
solute carrier family 38, member 6 |
| chr9_-_111696340 | 0.13 |
ENST00000374647.5
|
IKBKAP
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase complex-associated protein |
| chr19_+_2841433 | 0.13 |
ENST00000334241.4
ENST00000585966.1 ENST00000591539.1 |
ZNF555
|
zinc finger protein 555 |
| chr17_+_53342311 | 0.13 |
ENST00000226067.5
|
HLF
|
hepatic leukemia factor |
| chr19_+_36103631 | 0.13 |
ENST00000203166.5
ENST00000379045.2 |
HAUS5
|
HAUS augmin-like complex, subunit 5 |
| chr12_+_56511943 | 0.13 |
ENST00000257940.2
ENST00000552345.1 ENST00000551880.1 ENST00000546903.1 ENST00000551790.1 |
ZC3H10
ESYT1
|
zinc finger CCCH-type containing 10 extended synaptotagmin-like protein 1 |
| chr5_+_139055021 | 0.13 |
ENST00000502716.1
ENST00000503511.1 |
CXXC5
|
CXXC finger protein 5 |
| chr8_+_98788057 | 0.13 |
ENST00000517924.1
|
LAPTM4B
|
lysosomal protein transmembrane 4 beta |
| chr22_+_32340481 | 0.13 |
ENST00000397492.1
|
YWHAH
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta |
| chr12_-_31882108 | 0.13 |
ENST00000281471.6
|
AMN1
|
antagonist of mitotic exit network 1 homolog (S. cerevisiae) |
| chr19_+_36486078 | 0.13 |
ENST00000378887.2
|
SDHAF1
|
succinate dehydrogenase complex assembly factor 1 |
| chr2_-_216300784 | 0.13 |
ENST00000421182.1
ENST00000432072.2 ENST00000323926.6 ENST00000336916.4 ENST00000357867.4 ENST00000359671.1 ENST00000346544.3 ENST00000345488.5 ENST00000357009.2 ENST00000446046.1 ENST00000356005.4 ENST00000443816.1 ENST00000426059.1 ENST00000354785.4 |
FN1
|
fibronectin 1 |
| chr15_+_91498089 | 0.13 |
ENST00000394258.2
ENST00000555155.1 |
RCCD1
|
RCC1 domain containing 1 |
| chr19_-_55574538 | 0.12 |
ENST00000415061.3
|
RDH13
|
retinol dehydrogenase 13 (all-trans/9-cis) |
| chr11_+_18343800 | 0.12 |
ENST00000453096.2
|
GTF2H1
|
general transcription factor IIH, polypeptide 1, 62kDa |
| chr20_+_5892037 | 0.12 |
ENST00000378961.4
|
CHGB
|
chromogranin B (secretogranin 1) |
| chr7_-_45026419 | 0.12 |
ENST00000578968.1
ENST00000580528.1 |
SNHG15
|
small nucleolar RNA host gene 15 (non-protein coding) |
| chr12_-_12715266 | 0.12 |
ENST00000228862.2
|
DUSP16
|
dual specificity phosphatase 16 |
| chr14_+_100485712 | 0.12 |
ENST00000544450.2
|
EVL
|
Enah/Vasp-like |
| chr12_+_10365404 | 0.11 |
ENST00000266458.5
ENST00000421801.2 ENST00000544284.1 ENST00000545047.1 ENST00000543602.1 ENST00000545887.1 |
GABARAPL1
|
GABA(A) receptor-associated protein like 1 |
| chr7_+_23221613 | 0.11 |
ENST00000410002.3
ENST00000413919.1 |
NUPL2
|
nucleoporin like 2 |
| chr16_-_4588762 | 0.11 |
ENST00000562334.1
ENST00000562579.1 ENST00000567695.1 ENST00000563507.1 |
CDIP1
|
cell death-inducing p53 target 1 |
| chr19_+_10362882 | 0.11 |
ENST00000393733.2
ENST00000588502.1 |
MRPL4
|
mitochondrial ribosomal protein L4 |
| chr14_-_93651186 | 0.11 |
ENST00000556883.1
ENST00000298894.4 |
MOAP1
|
modulator of apoptosis 1 |
| chr12_-_100660833 | 0.11 |
ENST00000551642.1
ENST00000416321.1 ENST00000550587.1 ENST00000549249.1 |
DEPDC4
|
DEP domain containing 4 |
| chr20_+_17949724 | 0.11 |
ENST00000377704.4
|
MGME1
|
mitochondrial genome maintenance exonuclease 1 |
| chr12_+_54378923 | 0.11 |
ENST00000303460.4
|
HOXC10
|
homeobox C10 |
| chr3_-_154042205 | 0.11 |
ENST00000329463.5
|
DHX36
|
DEAH (Asp-Glu-Ala-His) box polypeptide 36 |
| chrX_-_152245978 | 0.11 |
ENST00000538162.2
|
PNMA6D
|
paraneoplastic Ma antigen family member 6D (pseudogene) |
| chr6_+_37225540 | 0.11 |
ENST00000373491.3
|
TBC1D22B
|
TBC1 domain family, member 22B |
| chr5_+_70751442 | 0.11 |
ENST00000358731.4
ENST00000380675.2 |
BDP1
|
B double prime 1, subunit of RNA polymerase III transcription initiation factor IIIB |
| chr4_+_108910870 | 0.11 |
ENST00000403312.1
ENST00000603302.1 ENST00000309522.3 |
HADH
|
hydroxyacyl-CoA dehydrogenase |
| chr6_-_30523865 | 0.11 |
ENST00000433809.1
|
GNL1
|
guanine nucleotide binding protein-like 1 |
| chr12_+_112856690 | 0.10 |
ENST00000392597.1
ENST00000351677.2 |
PTPN11
|
protein tyrosine phosphatase, non-receptor type 11 |
| chr16_-_86588627 | 0.10 |
ENST00000565482.1
ENST00000564364.1 ENST00000561989.1 ENST00000543303.2 ENST00000381214.5 ENST00000360900.6 ENST00000322911.6 ENST00000546093.1 ENST00000569000.1 ENST00000562994.1 ENST00000561522.1 |
MTHFSD
|
methenyltetrahydrofolate synthetase domain containing |
| chr1_+_28099683 | 0.10 |
ENST00000373943.4
|
STX12
|
syntaxin 12 |
| chr4_+_668348 | 0.10 |
ENST00000511290.1
|
MYL5
|
myosin, light chain 5, regulatory |
| chr12_-_31882027 | 0.10 |
ENST00000541931.1
ENST00000535408.1 |
AMN1
|
antagonist of mitotic exit network 1 homolog (S. cerevisiae) |
| chr15_+_42841008 | 0.10 |
ENST00000260372.3
ENST00000568876.1 ENST00000568846.2 ENST00000562398.1 |
HAUS2
|
HAUS augmin-like complex, subunit 2 |
| chr8_-_21966893 | 0.10 |
ENST00000522405.1
ENST00000522379.1 ENST00000309188.6 ENST00000521807.2 |
NUDT18
|
nudix (nucleoside diphosphate linked moiety X)-type motif 18 |
| chr11_+_809961 | 0.10 |
ENST00000530797.1
|
RPLP2
|
ribosomal protein, large, P2 |
| chr12_-_58165870 | 0.10 |
ENST00000257848.7
|
METTL1
|
methyltransferase like 1 |
| chr5_+_56469843 | 0.10 |
ENST00000514387.2
|
GPBP1
|
GC-rich promoter binding protein 1 |
| chr14_+_60716159 | 0.10 |
ENST00000325658.3
|
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent, 1A |
| chr3_+_44803322 | 0.10 |
ENST00000481166.2
|
KIF15
|
kinesin family member 15 |
| chr2_+_70142232 | 0.10 |
ENST00000540449.1
|
MXD1
|
MAX dimerization protein 1 |
| chr11_-_118122996 | 0.10 |
ENST00000525386.1
ENST00000527472.1 ENST00000278949.4 |
MPZL3
|
myelin protein zero-like 3 |
| chr17_-_41322332 | 0.10 |
ENST00000590740.1
|
RP11-242D8.1
|
RP11-242D8.1 |
| chr9_+_116298778 | 0.09 |
ENST00000462143.1
|
RGS3
|
regulator of G-protein signaling 3 |
| chr17_+_74536115 | 0.09 |
ENST00000592014.1
|
PRCD
|
progressive rod-cone degeneration |
| chr11_-_62389449 | 0.09 |
ENST00000534026.1
|
B3GAT3
|
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) |
| chr12_+_56522001 | 0.09 |
ENST00000267113.4
ENST00000541590.1 |
ESYT1
|
extended synaptotagmin-like protein 1 |
| chr18_-_12656715 | 0.09 |
ENST00000462226.1
ENST00000497844.2 ENST00000309836.5 ENST00000453447.2 |
SPIRE1
|
spire-type actin nucleation factor 1 |
| chr15_-_66790146 | 0.09 |
ENST00000316634.5
|
SNAPC5
|
small nuclear RNA activating complex, polypeptide 5, 19kDa |
| chr9_-_77643189 | 0.09 |
ENST00000376837.3
|
C9orf41
|
chromosome 9 open reading frame 41 |
| chrX_+_152338301 | 0.09 |
ENST00000453825.2
|
PNMA6A
|
paraneoplastic Ma antigen family member 6A |
| chr7_+_89975979 | 0.09 |
ENST00000257659.8
ENST00000222511.6 ENST00000417207.1 |
GTPBP10
|
GTP-binding protein 10 (putative) |
| chr19_-_40919271 | 0.09 |
ENST00000291825.7
ENST00000324001.7 |
PRX
|
periaxin |
| chr4_-_170533723 | 0.09 |
ENST00000510533.1
ENST00000439128.2 ENST00000511633.1 ENST00000512193.1 ENST00000507142.1 |
NEK1
|
NIMA-related kinase 1 |
| chr9_-_2844058 | 0.09 |
ENST00000397885.2
|
KIAA0020
|
KIAA0020 |
| chr2_-_224467002 | 0.09 |
ENST00000421386.1
ENST00000433889.1 |
SCG2
|
secretogranin II |
| chr19_+_49122548 | 0.08 |
ENST00000245222.4
ENST00000340932.3 ENST00000601712.1 ENST00000600537.1 |
SPHK2
|
sphingosine kinase 2 |
| chr7_+_23221438 | 0.08 |
ENST00000258742.5
|
NUPL2
|
nucleoporin like 2 |
| chr2_-_242255117 | 0.08 |
ENST00000420451.1
ENST00000417540.1 ENST00000310931.4 |
HDLBP
|
high density lipoprotein binding protein |
| chr17_-_49124230 | 0.08 |
ENST00000510283.1
ENST00000510855.1 |
SPAG9
|
sperm associated antigen 9 |
| chr15_+_84841242 | 0.08 |
ENST00000558195.1
|
RP11-182J1.16
|
ubiquitin-conjugating enzyme E2Q family member 2-like |
| chr5_-_145562147 | 0.08 |
ENST00000545646.1
ENST00000274562.9 ENST00000510191.1 ENST00000394434.2 |
LARS
|
leucyl-tRNA synthetase |
| chr16_+_27413483 | 0.08 |
ENST00000337929.3
ENST00000564089.1 |
IL21R
|
interleukin 21 receptor |
| chr15_-_66790047 | 0.08 |
ENST00000566658.1
ENST00000563480.2 ENST00000395589.2 ENST00000307979.7 |
SNAPC5
|
small nuclear RNA activating complex, polypeptide 5, 19kDa |
| chr3_+_101292939 | 0.08 |
ENST00000265260.3
ENST00000469941.1 ENST00000296024.5 |
PCNP
|
PEST proteolytic signal containing nuclear protein |
| chr2_-_21022818 | 0.08 |
ENST00000440866.2
ENST00000541941.1 ENST00000402479.2 ENST00000435420.2 ENST00000432947.1 ENST00000403006.2 ENST00000419825.2 ENST00000381090.3 ENST00000237822.3 ENST00000412261.1 |
C2orf43
|
chromosome 2 open reading frame 43 |
| chr7_-_140624499 | 0.08 |
ENST00000288602.6
|
BRAF
|
v-raf murine sarcoma viral oncogene homolog B |
| chr14_+_60715928 | 0.08 |
ENST00000395076.4
|
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent, 1A |
| chr10_+_123923205 | 0.08 |
ENST00000369004.3
ENST00000260733.3 |
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
| chr15_+_96904487 | 0.07 |
ENST00000600790.1
|
AC087477.1
|
Uncharacterized protein |
| chr10_+_119000604 | 0.07 |
ENST00000298472.5
|
SLC18A2
|
solute carrier family 18 (vesicular monoamine transporter), member 2 |
| chr11_-_3818688 | 0.07 |
ENST00000355260.3
ENST00000397004.4 ENST00000397007.4 ENST00000532475.1 |
NUP98
|
nucleoporin 98kDa |
| chr8_-_10697370 | 0.07 |
ENST00000314787.3
ENST00000426190.2 ENST00000519088.1 |
PINX1
|
PIN2/TERF1 interacting, telomerase inhibitor 1 |
| chr9_+_131644398 | 0.07 |
ENST00000372599.3
|
LRRC8A
|
leucine rich repeat containing 8 family, member A |
| chr9_+_131644781 | 0.07 |
ENST00000259324.5
|
LRRC8A
|
leucine rich repeat containing 8 family, member A |
| chr16_-_27561209 | 0.07 |
ENST00000356183.4
ENST00000561623.1 |
GTF3C1
|
general transcription factor IIIC, polypeptide 1, alpha 220kDa |
| chr5_-_95297678 | 0.07 |
ENST00000237853.4
|
ELL2
|
elongation factor, RNA polymerase II, 2 |
| chr4_+_128802016 | 0.07 |
ENST00000270861.5
ENST00000515069.1 ENST00000513090.1 ENST00000507249.1 |
PLK4
|
polo-like kinase 4 |
| chr4_+_667686 | 0.07 |
ENST00000505477.1
|
MYL5
|
myosin, light chain 5, regulatory |
| chr2_-_43823119 | 0.07 |
ENST00000403856.1
ENST00000404790.1 ENST00000405975.2 ENST00000415080.2 |
THADA
|
thyroid adenoma associated |
| chr17_-_43210580 | 0.07 |
ENST00000538093.1
ENST00000590644.1 |
PLCD3
|
phospholipase C, delta 3 |
| chr7_+_99905325 | 0.07 |
ENST00000332397.6
ENST00000437326.2 |
SPDYE3
|
speedy/RINGO cell cycle regulator family member E3 |
| chr2_+_220071490 | 0.07 |
ENST00000409206.1
ENST00000409594.1 ENST00000289528.5 ENST00000422255.1 ENST00000409412.1 ENST00000409097.1 ENST00000409336.1 ENST00000409217.1 ENST00000409319.1 ENST00000444522.2 |
ZFAND2B
|
zinc finger, AN1-type domain 2B |
| chr13_+_111767650 | 0.07 |
ENST00000449979.1
ENST00000370623.3 |
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr9_+_97488939 | 0.07 |
ENST00000277198.2
ENST00000297979.5 |
C9orf3
|
chromosome 9 open reading frame 3 |
| chr12_+_7013897 | 0.07 |
ENST00000007969.8
ENST00000323702.5 |
LRRC23
|
leucine rich repeat containing 23 |
| chr7_+_140396465 | 0.07 |
ENST00000476279.1
ENST00000247866.4 ENST00000461457.1 ENST00000465506.1 ENST00000204307.5 ENST00000464566.1 |
NDUFB2
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2, 8kDa |
| chr5_+_56469939 | 0.07 |
ENST00000506184.2
|
GPBP1
|
GC-rich promoter binding protein 1 |
| chr1_+_29213584 | 0.07 |
ENST00000343067.4
ENST00000356093.2 ENST00000398863.2 ENST00000373800.3 ENST00000349460.4 |
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) |
| chr11_-_62389621 | 0.07 |
ENST00000531383.1
ENST00000265471.5 |
B3GAT3
|
beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) |
| chr1_+_6684918 | 0.07 |
ENST00000054650.4
|
THAP3
|
THAP domain containing, apoptosis associated protein 3 |
| chr22_+_41253080 | 0.07 |
ENST00000541156.1
ENST00000414396.1 ENST00000357137.4 |
XPNPEP3
|
X-prolyl aminopeptidase (aminopeptidase P) 3, putative |
| chr12_-_56694083 | 0.07 |
ENST00000552688.1
ENST00000548041.1 ENST00000551137.1 ENST00000551968.1 ENST00000542324.2 ENST00000546930.1 ENST00000549221.1 ENST00000550159.1 ENST00000550734.1 |
CS
|
citrate synthase |
| chr9_-_125027079 | 0.07 |
ENST00000417201.3
|
RBM18
|
RNA binding motif protein 18 |
| chr2_+_108994466 | 0.07 |
ENST00000272452.2
|
SULT1C4
|
sulfotransferase family, cytosolic, 1C, member 4 |
| chr4_-_77997126 | 0.07 |
ENST00000537948.1
ENST00000507788.1 ENST00000237654.4 |
CCNI
|
cyclin I |
| chr12_+_122516626 | 0.07 |
ENST00000319080.7
|
MLXIP
|
MLX interacting protein |
| chr16_+_19079311 | 0.06 |
ENST00000569127.1
|
COQ7
|
coenzyme Q7 homolog, ubiquinone (yeast) |
| chr19_+_10362577 | 0.06 |
ENST00000592514.1
ENST00000307422.5 ENST00000253099.6 ENST00000590150.1 ENST00000590669.1 |
MRPL4
|
mitochondrial ribosomal protein L4 |
| chr4_-_39367949 | 0.06 |
ENST00000503784.1
ENST00000349703.2 ENST00000381897.1 |
RFC1
|
replication factor C (activator 1) 1, 145kDa |
| chr18_+_72163443 | 0.06 |
ENST00000324262.4
ENST00000580672.1 |
CNDP2
|
CNDP dipeptidase 2 (metallopeptidase M20 family) |
| chr12_+_100661156 | 0.06 |
ENST00000360820.2
|
SCYL2
|
SCY1-like 2 (S. cerevisiae) |
| chr21_-_44846999 | 0.06 |
ENST00000270162.6
|
SIK1
|
salt-inducible kinase 1 |
| chr10_+_123922941 | 0.06 |
ENST00000360561.3
|
TACC2
|
transforming, acidic coiled-coil containing protein 2 |
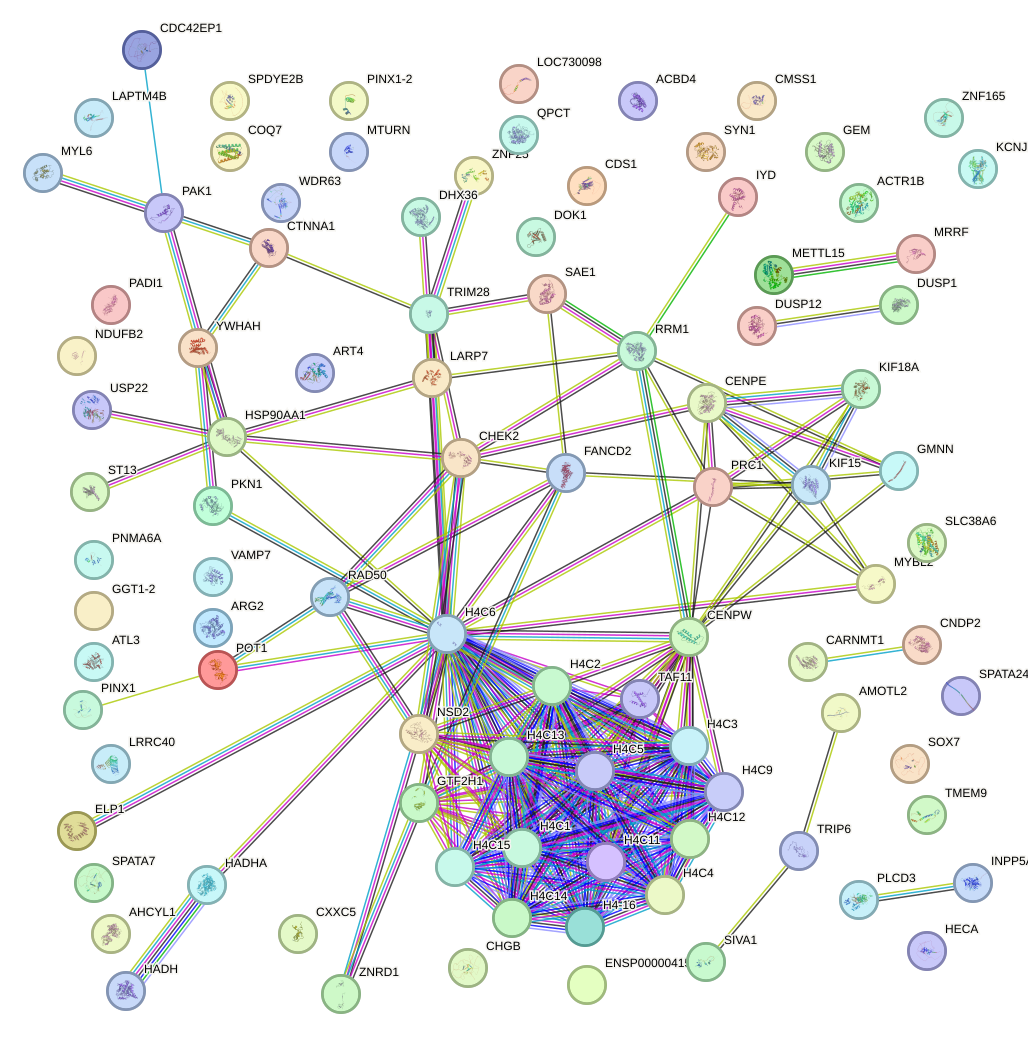
Network of associatons between targets according to the STRING database.
First level regulatory network of ATF7
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0071373 | cellular response to luteinizing hormone stimulus(GO:0071373) |
| 0.1 | 0.3 | GO:1901536 | negative regulation of DNA demethylation(GO:1901536) |
| 0.1 | 0.2 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.2 | GO:0060382 | regulation of DNA strand elongation(GO:0060382) |
| 0.1 | 0.3 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.1 | 0.2 | GO:0017186 | peptidyl-pyroglutamic acid biosynthetic process, using glutaminyl-peptide cyclotransferase(GO:0017186) |
| 0.0 | 0.1 | GO:1903595 | positive regulation of histamine secretion by mast cell(GO:1903595) |
| 0.0 | 0.3 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.0 | 0.2 | GO:1904744 | positive regulation of telomeric DNA binding(GO:1904744) |
| 0.0 | 0.1 | GO:0034553 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.1 | GO:1904237 | regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904235) positive regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904237) |
| 0.0 | 0.1 | GO:1901895 | negative regulation of calcium-transporting ATPase activity(GO:1901895) |
| 0.0 | 0.3 | GO:1902739 | interferon-alpha secretion(GO:0072642) telomerase RNA stabilization(GO:0090669) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 0.0 | 0.2 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.2 | GO:2001045 | negative regulation of integrin-mediated signaling pathway(GO:2001045) |
| 0.0 | 0.1 | GO:0070898 | RNA polymerase III transcriptional preinitiation complex assembly(GO:0070898) |
| 0.0 | 0.1 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.0 | 0.2 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.2 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.0 | 0.1 | GO:0045208 | MAPK phosphatase export from nucleus(GO:0045208) MAPK phosphatase export from nucleus, leptomycin B sensitive(GO:0045209) |
| 0.0 | 0.2 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.2 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.1 | GO:0006429 | glutaminyl-tRNA aminoacylation(GO:0006425) leucyl-tRNA aminoacylation(GO:0006429) |
| 0.0 | 0.2 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.2 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 | 0.1 | GO:0061582 | intestinal epithelial cell migration(GO:0061582) |
| 0.0 | 0.3 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.2 | GO:0061084 | regulation of protein refolding(GO:0061083) negative regulation of protein refolding(GO:0061084) |
| 0.0 | 0.1 | GO:0042137 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) sequestering of neurotransmitter(GO:0042137) |
| 0.0 | 0.4 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.2 | GO:0060244 | negative regulation of cell proliferation involved in contact inhibition(GO:0060244) |
| 0.0 | 0.2 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.2 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.3 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.1 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.0 | 0.2 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.2 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.2 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.2 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.3 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.0 | 0.3 | GO:0046855 | inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.2 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.2 | GO:0000050 | urea cycle(GO:0000050) |
| 0.0 | 0.5 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.2 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 0.0 | 0.1 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.0 | 0.3 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 0.1 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.1 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.0 | 0.1 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.0 | 0.0 | GO:2000681 | negative regulation of rubidium ion transport(GO:2000681) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.0 | 0.1 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.3 | GO:0006206 | pyrimidine nucleobase metabolic process(GO:0006206) |
| 0.0 | 0.0 | GO:0019240 | citrulline biosynthetic process(GO:0019240) |
| 0.0 | 0.1 | GO:0070649 | polar body extrusion after meiotic divisions(GO:0040038) formin-nucleated actin cable assembly(GO:0070649) |
| 0.0 | 0.3 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.3 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.3 | GO:0060716 | labyrinthine layer blood vessel development(GO:0060716) |
| 0.0 | 0.1 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.1 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.1 | 0.3 | GO:0005873 | plus-end kinesin complex(GO:0005873) |
| 0.1 | 0.4 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.2 | GO:0031523 | Myb complex(GO:0031523) |
| 0.0 | 0.3 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.3 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.5 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.3 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.1 | GO:0000126 | transcription factor TFIIIB complex(GO:0000126) |
| 0.0 | 0.3 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.4 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.4 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.2 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.2 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.2 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.1 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.1 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 0.3 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.5 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.2 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.2 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.1 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.2 | GO:0016600 | flotillin complex(GO:0016600) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0019948 | SUMO activating enzyme activity(GO:0019948) |
| 0.1 | 0.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.1 | 0.4 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.1 | 0.2 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.1 | 0.2 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.1 | 0.2 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.2 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.0 | 0.2 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.0 | 0.3 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.1 | GO:0004108 | citrate (Si)-synthase activity(GO:0004108) citrate synthase activity(GO:0036440) |
| 0.0 | 0.1 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.0 | 0.1 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.0 | 0.2 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.5 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.3 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.2 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.3 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.3 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.1 | GO:0004823 | glutamine-tRNA ligase activity(GO:0004819) leucine-tRNA ligase activity(GO:0004823) |
| 0.0 | 0.5 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.1 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.0 | 0.2 | GO:0032558 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.0 | 0.1 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 0.1 | GO:0001003 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) |
| 0.0 | 0.2 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.3 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) |
| 0.0 | 0.5 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.5 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.2 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.4 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.3 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.2 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.2 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.0 | 0.2 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.3 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.3 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.1 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.2 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.5 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.2 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.2 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |