Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
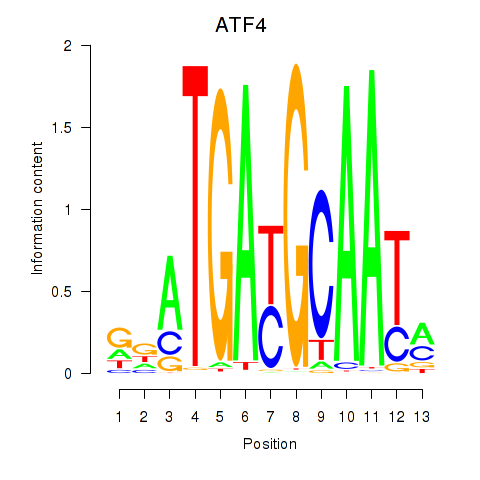
Results for ATF4
Z-value: 0.30
Transcription factors associated with ATF4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ATF4
|
ENSG00000128272.10 | activating transcription factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ATF4 | hg19_v2_chr22_+_39916558_39916579 | 0.77 | 2.3e-01 | Click! |
Activity profile of ATF4 motif
Sorted Z-values of ATF4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_41245160 | 0.55 |
ENST00000444189.2
ENST00000446533.3 |
CHAC1
|
ChaC, cation transport regulator homolog 1 (E. coli) |
| chr15_+_89182178 | 0.26 |
ENST00000559876.1
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr15_+_89182156 | 0.26 |
ENST00000379224.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chrX_-_133792480 | 0.22 |
ENST00000359237.4
|
PLAC1
|
placenta-specific 1 |
| chr11_-_33913708 | 0.21 |
ENST00000257818.2
|
LMO2
|
LIM domain only 2 (rhombotin-like 1) |
| chr15_+_89181974 | 0.21 |
ENST00000306072.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr19_+_11350278 | 0.19 |
ENST00000252453.8
|
C19orf80
|
chromosome 19 open reading frame 80 |
| chr2_+_65663812 | 0.18 |
ENST00000606978.1
ENST00000377977.3 ENST00000536804.1 |
AC074391.1
|
AC074391.1 |
| chr7_-_99006443 | 0.15 |
ENST00000350498.3
|
PDAP1
|
PDGFA associated protein 1 |
| chr10_+_102106829 | 0.13 |
ENST00000370355.2
|
SCD
|
stearoyl-CoA desaturase (delta-9-desaturase) |
| chr1_-_149832704 | 0.13 |
ENST00000392933.1
ENST00000369157.2 ENST00000392932.4 |
HIST2H4B
|
histone cluster 2, H4b |
| chr3_+_11314099 | 0.12 |
ENST00000446450.2
ENST00000354956.5 ENST00000354449.3 ENST00000419112.1 |
ATG7
|
autophagy related 7 |
| chr12_+_93963590 | 0.12 |
ENST00000340600.2
|
SOCS2
|
suppressor of cytokine signaling 2 |
| chr7_+_99006550 | 0.12 |
ENST00000222969.5
|
BUD31
|
BUD31 homolog (S. cerevisiae) |
| chr7_-_48068699 | 0.12 |
ENST00000412142.1
ENST00000395572.2 |
SUN3
|
Sad1 and UNC84 domain containing 3 |
| chr7_-_48068671 | 0.11 |
ENST00000297325.4
|
SUN3
|
Sad1 and UNC84 domain containing 3 |
| chr11_+_58938903 | 0.11 |
ENST00000532982.1
|
DTX4
|
deltex homolog 4 (Drosophila) |
| chr1_-_44497118 | 0.11 |
ENST00000537678.1
ENST00000466926.1 |
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr1_-_178840157 | 0.10 |
ENST00000367629.1
ENST00000234816.2 |
ANGPTL1
|
angiopoietin-like 1 |
| chr5_+_143584814 | 0.10 |
ENST00000507359.3
|
KCTD16
|
potassium channel tetramerization domain containing 16 |
| chr16_-_67694597 | 0.10 |
ENST00000393919.4
ENST00000219251.8 |
ACD
|
adrenocortical dysplasia homolog (mouse) |
| chr11_-_3078616 | 0.09 |
ENST00000401769.3
ENST00000278224.9 ENST00000397114.3 ENST00000380525.4 |
CARS
|
cysteinyl-tRNA synthetase |
| chr11_+_93754513 | 0.09 |
ENST00000315765.9
|
HEPHL1
|
hephaestin-like 1 |
| chr6_+_126102292 | 0.09 |
ENST00000368357.3
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr7_+_99006232 | 0.08 |
ENST00000403633.2
|
BUD31
|
BUD31 homolog (S. cerevisiae) |
| chrX_-_71792477 | 0.08 |
ENST00000421523.1
ENST00000415409.1 ENST00000373559.4 ENST00000373556.3 ENST00000373560.2 ENST00000373583.1 ENST00000429103.2 ENST00000373571.1 ENST00000373554.1 |
HDAC8
|
histone deacetylase 8 |
| chr5_+_167956121 | 0.08 |
ENST00000338333.4
|
FBLL1
|
fibrillarin-like 1 |
| chr6_+_151187074 | 0.08 |
ENST00000367308.4
|
MTHFD1L
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like |
| chr11_-_3078838 | 0.07 |
ENST00000397111.5
|
CARS
|
cysteinyl-tRNA synthetase |
| chr12_+_64798826 | 0.07 |
ENST00000540203.1
|
XPOT
|
exportin, tRNA |
| chr3_-_99595037 | 0.07 |
ENST00000383694.2
|
FILIP1L
|
filamin A interacting protein 1-like |
| chrX_+_77359726 | 0.07 |
ENST00000442431.1
|
PGK1
|
phosphoglycerate kinase 1 |
| chr1_+_154244987 | 0.06 |
ENST00000328703.7
ENST00000457918.2 ENST00000483970.2 ENST00000435087.1 ENST00000532105.1 |
HAX1
|
HCLS1 associated protein X-1 |
| chr1_+_241815577 | 0.06 |
ENST00000366552.2
ENST00000437684.2 |
WDR64
|
WD repeat domain 64 |
| chr1_-_44497024 | 0.06 |
ENST00000372306.3
ENST00000372310.3 ENST00000475075.2 |
SLC6A9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr14_-_23762777 | 0.06 |
ENST00000431326.2
|
HOMEZ
|
homeobox and leucine zipper encoding |
| chr9_-_34665983 | 0.06 |
ENST00000416454.1
ENST00000544078.2 ENST00000421828.2 ENST00000423809.1 |
RP11-195F19.5
|
HCG2040265, isoform CRA_a; Uncharacterized protein; cDNA FLJ50015 |
| chr1_+_149804218 | 0.06 |
ENST00000610125.1
|
HIST2H4A
|
histone cluster 2, H4a |
| chr14_-_88282595 | 0.06 |
ENST00000554519.1
|
RP11-1152H15.1
|
RP11-1152H15.1 |
| chr3_-_99594948 | 0.06 |
ENST00000471562.1
ENST00000495625.2 |
FILIP1L
|
filamin A interacting protein 1-like |
| chr11_+_118955583 | 0.06 |
ENST00000278715.3
ENST00000536813.1 ENST00000537841.1 ENST00000542729.1 ENST00000546302.1 ENST00000442944.2 ENST00000544387.1 ENST00000543090.1 |
HMBS
|
hydroxymethylbilane synthase |
| chr5_+_174151536 | 0.06 |
ENST00000239243.6
ENST00000507785.1 |
MSX2
|
msh homeobox 2 |
| chr12_-_25101920 | 0.06 |
ENST00000539780.1
ENST00000546285.1 ENST00000342945.5 |
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr22_-_29457832 | 0.06 |
ENST00000216071.4
|
C22orf31
|
chromosome 22 open reading frame 31 |
| chr3_-_48130707 | 0.06 |
ENST00000360240.6
ENST00000383737.4 |
MAP4
|
microtubule-associated protein 4 |
| chr17_-_77967433 | 0.05 |
ENST00000571872.1
|
TBC1D16
|
TBC1 domain family, member 16 |
| chr1_+_162351503 | 0.05 |
ENST00000458626.2
|
C1orf226
|
chromosome 1 open reading frame 226 |
| chrX_+_77359671 | 0.05 |
ENST00000373316.4
|
PGK1
|
phosphoglycerate kinase 1 |
| chr17_+_73089382 | 0.05 |
ENST00000538213.2
ENST00000584118.1 |
SLC16A5
|
solute carrier family 16 (monocarboxylate transporter), member 5 |
| chr1_+_239882842 | 0.05 |
ENST00000448020.1
|
CHRM3
|
cholinergic receptor, muscarinic 3 |
| chr7_-_50860565 | 0.05 |
ENST00000403097.1
|
GRB10
|
growth factor receptor-bound protein 10 |
| chr12_+_57914742 | 0.05 |
ENST00000551351.1
|
MBD6
|
methyl-CpG binding domain protein 6 |
| chr12_-_25102252 | 0.05 |
ENST00000261192.7
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr8_-_116681686 | 0.05 |
ENST00000519815.1
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr1_+_112016414 | 0.05 |
ENST00000343534.5
ENST00000369718.3 |
C1orf162
|
chromosome 1 open reading frame 162 |
| chr8_-_38008783 | 0.04 |
ENST00000276449.4
|
STAR
|
steroidogenic acute regulatory protein |
| chr12_+_56862301 | 0.04 |
ENST00000338146.5
|
SPRYD4
|
SPRY domain containing 4 |
| chr17_-_77813186 | 0.04 |
ENST00000448310.1
ENST00000269397.4 |
CBX4
|
chromobox homolog 4 |
| chr1_+_52521928 | 0.04 |
ENST00000489308.2
|
BTF3L4
|
basic transcription factor 3-like 4 |
| chr11_+_62649158 | 0.04 |
ENST00000539891.1
ENST00000536981.1 |
SLC3A2
|
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr12_+_106994905 | 0.04 |
ENST00000357881.4
|
RFX4
|
regulatory factor X, 4 (influences HLA class II expression) |
| chrX_-_15332665 | 0.04 |
ENST00000537676.1
ENST00000344384.4 |
ASB11
|
ankyrin repeat and SOCS box containing 11 |
| chr11_-_128737163 | 0.04 |
ENST00000324003.3
ENST00000392665.2 |
KCNJ1
|
potassium inwardly-rectifying channel, subfamily J, member 1 |
| chr4_-_70725856 | 0.04 |
ENST00000226444.3
|
SULT1E1
|
sulfotransferase family 1E, estrogen-preferring, member 1 |
| chr16_-_70323422 | 0.04 |
ENST00000261772.8
|
AARS
|
alanyl-tRNA synthetase |
| chr5_+_154135029 | 0.04 |
ENST00000518297.1
|
LARP1
|
La ribonucleoprotein domain family, member 1 |
| chr19_-_51014460 | 0.04 |
ENST00000595669.1
|
JOSD2
|
Josephin domain containing 2 |
| chr12_+_57623907 | 0.04 |
ENST00000553529.1
ENST00000554310.1 |
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr7_+_30634297 | 0.04 |
ENST00000389266.3
|
GARS
|
glycyl-tRNA synthetase |
| chr14_+_75894714 | 0.04 |
ENST00000559060.1
|
JDP2
|
Jun dimerization protein 2 |
| chr10_+_50507232 | 0.04 |
ENST00000374144.3
|
C10orf71
|
chromosome 10 open reading frame 71 |
| chr7_-_93633684 | 0.03 |
ENST00000222547.3
ENST00000425626.1 |
BET1
|
Bet1 golgi vesicular membrane trafficking protein |
| chr7_-_115670792 | 0.03 |
ENST00000265440.7
ENST00000393485.1 |
TFEC
|
transcription factor EC |
| chr3_+_11314072 | 0.03 |
ENST00000444619.1
|
ATG7
|
autophagy related 7 |
| chr17_-_76836729 | 0.03 |
ENST00000587783.1
ENST00000542802.3 ENST00000586531.1 ENST00000589424.1 ENST00000590546.2 |
USP36
|
ubiquitin specific peptidase 36 |
| chr15_+_40886439 | 0.03 |
ENST00000532056.1
ENST00000399668.2 |
CASC5
|
cancer susceptibility candidate 5 |
| chr1_-_216978709 | 0.03 |
ENST00000360012.3
|
ESRRG
|
estrogen-related receptor gamma |
| chr8_-_116680833 | 0.03 |
ENST00000220888.5
|
TRPS1
|
trichorhinophalangeal syndrome I |
| chr11_-_128737259 | 0.03 |
ENST00000440599.2
ENST00000392666.1 ENST00000324036.3 |
KCNJ1
|
potassium inwardly-rectifying channel, subfamily J, member 1 |
| chr17_-_76836963 | 0.03 |
ENST00000312010.6
|
USP36
|
ubiquitin specific peptidase 36 |
| chr19_-_51014588 | 0.03 |
ENST00000598418.1
|
JOSD2
|
Josephin domain containing 2 |
| chr7_-_115670804 | 0.02 |
ENST00000320239.7
|
TFEC
|
transcription factor EC |
| chr22_-_43485381 | 0.02 |
ENST00000331018.7
ENST00000266254.7 ENST00000445824.1 |
TTLL1
|
tubulin tyrosine ligase-like family, member 1 |
| chr22_+_50919944 | 0.02 |
ENST00000395738.2
|
ADM2
|
adrenomedullin 2 |
| chr12_+_57914481 | 0.02 |
ENST00000548887.1
|
MBD6
|
methyl-CpG binding domain protein 6 |
| chr3_-_48130314 | 0.02 |
ENST00000439356.1
ENST00000395734.3 ENST00000426837.2 |
MAP4
|
microtubule-associated protein 4 |
| chr22_+_42229100 | 0.02 |
ENST00000361204.4
|
SREBF2
|
sterol regulatory element binding transcription factor 2 |
| chr7_-_138386097 | 0.02 |
ENST00000421622.1
|
SVOPL
|
SVOP-like |
| chrX_+_48398053 | 0.02 |
ENST00000537536.1
ENST00000418627.1 |
TBC1D25
|
TBC1 domain family, member 25 |
| chr17_-_79895097 | 0.02 |
ENST00000402252.2
ENST00000583564.1 ENST00000585244.1 ENST00000337943.5 ENST00000579698.1 |
PYCR1
|
pyrroline-5-carboxylate reductase 1 |
| chr12_+_57623869 | 0.02 |
ENST00000414700.3
ENST00000557703.1 |
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr1_+_162602244 | 0.02 |
ENST00000367922.3
ENST00000367921.3 |
DDR2
|
discoidin domain receptor tyrosine kinase 2 |
| chr19_-_51014345 | 0.02 |
ENST00000391815.3
ENST00000594350.1 ENST00000601423.1 |
JOSD2
|
Josephin domain containing 2 |
| chr1_-_167883353 | 0.02 |
ENST00000545172.1
|
ADCY10
|
adenylate cyclase 10 (soluble) |
| chr17_-_79895154 | 0.02 |
ENST00000405481.4
ENST00000585215.1 ENST00000577624.1 ENST00000403172.4 |
PYCR1
|
pyrroline-5-carboxylate reductase 1 |
| chr7_-_48068643 | 0.02 |
ENST00000453192.2
|
SUN3
|
Sad1 and UNC84 domain containing 3 |
| chr19_-_49258606 | 0.02 |
ENST00000310160.3
|
FUT1
|
fucosyltransferase 1 (galactoside 2-alpha-L-fucosyltransferase, H blood group) |
| chr2_-_98280383 | 0.02 |
ENST00000289228.5
|
ACTR1B
|
ARP1 actin-related protein 1 homolog B, centractin beta (yeast) |
| chrX_-_68385354 | 0.02 |
ENST00000361478.1
|
PJA1
|
praja ring finger 1, E3 ubiquitin protein ligase |
| chr4_-_153457197 | 0.02 |
ENST00000281708.4
|
FBXW7
|
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr9_+_36169380 | 0.02 |
ENST00000335119.2
|
CCIN
|
calicin |
| chr19_-_51192661 | 0.02 |
ENST00000391813.1
|
SHANK1
|
SH3 and multiple ankyrin repeat domains 1 |
| chr15_+_40886199 | 0.01 |
ENST00000346991.5
ENST00000528975.1 ENST00000527044.1 |
CASC5
|
cancer susceptibility candidate 5 |
| chr14_+_24563510 | 0.01 |
ENST00000545054.2
ENST00000561286.1 ENST00000558096.1 |
PCK2
|
phosphoenolpyruvate carboxykinase 2 (mitochondrial) |
| chr12_+_57624059 | 0.01 |
ENST00000557427.1
|
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr22_+_50919995 | 0.01 |
ENST00000362068.2
ENST00000395737.1 |
ADM2
|
adrenomedullin 2 |
| chr21_-_44495964 | 0.01 |
ENST00000398168.1
ENST00000398165.3 |
CBS
|
cystathionine-beta-synthase |
| chr3_-_33686743 | 0.01 |
ENST00000333778.6
ENST00000539981.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chrX_-_68385274 | 0.01 |
ENST00000374584.3
ENST00000590146.1 |
PJA1
|
praja ring finger 1, E3 ubiquitin protein ligase |
| chr12_-_10324716 | 0.01 |
ENST00000545927.1
ENST00000432556.2 ENST00000309539.3 ENST00000544577.1 |
OLR1
|
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chr3_-_33686925 | 0.01 |
ENST00000485378.2
ENST00000313350.6 ENST00000487200.1 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr2_+_7118755 | 0.01 |
ENST00000433456.1
|
RNF144A
|
ring finger protein 144A |
| chr12_+_6493406 | 0.01 |
ENST00000543190.1
|
LTBR
|
lymphotoxin beta receptor (TNFR superfamily, member 3) |
| chr12_+_57624119 | 0.01 |
ENST00000555773.1
ENST00000554975.1 ENST00000449049.3 ENST00000393827.4 |
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr7_+_48211048 | 0.01 |
ENST00000435803.1
|
ABCA13
|
ATP-binding cassette, sub-family A (ABC1), member 13 |
| chr12_+_57623477 | 0.01 |
ENST00000557487.1
ENST00000555634.1 ENST00000556689.1 |
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr12_+_50135588 | 0.01 |
ENST00000423828.1
ENST00000550445.1 |
TMBIM6
|
transmembrane BAX inhibitor motif containing 6 |
| chr12_+_57624085 | 0.00 |
ENST00000553474.1
|
SHMT2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr1_+_228353495 | 0.00 |
ENST00000366711.3
|
IBA57
|
IBA57, iron-sulfur cluster assembly homolog (S. cerevisiae) |
| chr5_+_33440802 | 0.00 |
ENST00000502553.1
ENST00000514259.1 ENST00000265112.3 |
TARS
|
threonyl-tRNA synthetase |
Network of associatons between targets according to the STRING database.
First level regulatory network of ATF4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.1 | 0.2 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.1 | 0.2 | GO:0006423 | cysteinyl-tRNA aminoacylation(GO:0006423) |
| 0.0 | 0.1 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.1 | GO:0060380 | regulation of single-stranded telomeric DNA binding(GO:0060380) positive regulation of single-stranded telomeric DNA binding(GO:0060381) |
| 0.0 | 0.1 | GO:0034727 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) suppression by virus of host autophagy(GO:0039521) |
| 0.0 | 0.1 | GO:0071922 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.5 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.0 | 0.1 | GO:0033967 | box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.0 | 0.2 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.0 | 0.1 | GO:0051795 | positive regulation of catagen(GO:0051795) |
| 0.0 | 0.1 | GO:0009082 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.0 | 0.0 | GO:0017143 | insecticide metabolic process(GO:0017143) cellular response to luteinizing hormone stimulus(GO:0071373) |
| 0.0 | 0.0 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.0 | 0.0 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.0 | GO:0032279 | asymmetric synapse(GO:0032279) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.1 | 0.5 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.1 | 0.2 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.1 | 0.2 | GO:0004817 | cysteine-tRNA ligase activity(GO:0004817) |
| 0.0 | 0.1 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.0 | 0.1 | GO:0019778 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.0 | 0.1 | GO:1990259 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.0 | 0.1 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.0 | 0.1 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.1 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.0 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |