Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
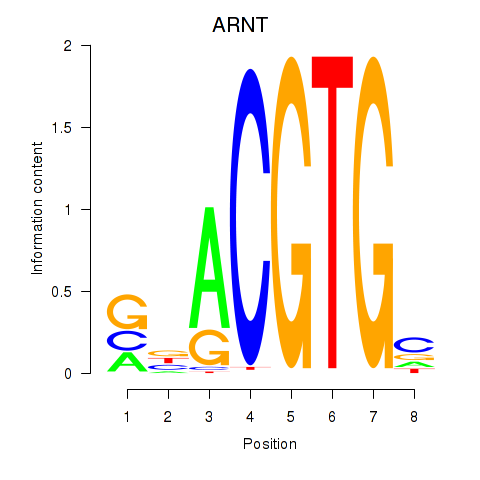
Results for ARNT
Z-value: 1.86
Transcription factors associated with ARNT
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ARNT
|
ENSG00000143437.16 | aryl hydrocarbon receptor nuclear translocator |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ARNT | hg19_v2_chr1_-_150849208_150849244 | -0.54 | 4.6e-01 | Click! |
Activity profile of ARNT motif
Sorted Z-values of ARNT motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_172755056 | 2.72 |
ENST00000520648.1
|
STC2
|
stanniocalcin 2 |
| chr15_+_45722727 | 2.22 |
ENST00000396650.2
ENST00000558435.1 ENST00000344300.3 |
C15orf48
|
chromosome 15 open reading frame 48 |
| chr5_-_131826457 | 1.65 |
ENST00000437654.1
ENST00000245414.4 |
IRF1
|
interferon regulatory factor 1 |
| chr1_-_8939265 | 1.51 |
ENST00000489867.1
|
ENO1
|
enolase 1, (alpha) |
| chr4_+_17578815 | 1.49 |
ENST00000226299.4
|
LAP3
|
leucine aminopeptidase 3 |
| chrX_+_77359726 | 1.40 |
ENST00000442431.1
|
PGK1
|
phosphoglycerate kinase 1 |
| chr3_-_36986534 | 1.40 |
ENST00000429976.2
ENST00000301807.6 |
TRANK1
|
tetratricopeptide repeat and ankyrin repeat containing 1 |
| chr2_+_176972000 | 1.37 |
ENST00000249504.5
|
HOXD11
|
homeobox D11 |
| chrX_+_77359671 | 1.29 |
ENST00000373316.4
|
PGK1
|
phosphoglycerate kinase 1 |
| chr3_+_38179969 | 1.24 |
ENST00000396334.3
ENST00000417037.2 ENST00000424893.1 ENST00000495303.1 ENST00000443433.2 ENST00000421516.1 |
MYD88
|
myeloid differentiation primary response 88 |
| chr22_-_42310570 | 1.18 |
ENST00000457093.1
|
SHISA8
|
shisa family member 8 |
| chr5_+_38846101 | 1.08 |
ENST00000274276.3
|
OSMR
|
oncostatin M receptor |
| chr19_-_45909585 | 1.08 |
ENST00000593226.1
ENST00000418234.2 |
PPP1R13L
|
protein phosphatase 1, regulatory subunit 13 like |
| chr13_-_44361025 | 1.01 |
ENST00000261488.6
|
ENOX1
|
ecto-NOX disulfide-thiol exchanger 1 |
| chr5_-_38845812 | 0.99 |
ENST00000513480.1
ENST00000512519.1 |
CTD-2127H9.1
|
CTD-2127H9.1 |
| chr11_+_57435441 | 0.96 |
ENST00000528177.1
|
ZDHHC5
|
zinc finger, DHHC-type containing 5 |
| chr6_-_139695757 | 0.96 |
ENST00000367651.2
|
CITED2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
| chr15_+_89182178 | 0.94 |
ENST00000559876.1
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr15_+_89182156 | 0.94 |
ENST00000379224.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr1_-_223537475 | 0.92 |
ENST00000344029.6
ENST00000494793.2 ENST00000366878.4 ENST00000366877.3 |
SUSD4
|
sushi domain containing 4 |
| chr11_+_72929319 | 0.89 |
ENST00000393597.2
ENST00000311131.2 |
P2RY2
|
purinergic receptor P2Y, G-protein coupled, 2 |
| chr16_+_3070313 | 0.86 |
ENST00000326577.4
|
TNFRSF12A
|
tumor necrosis factor receptor superfamily, member 12A |
| chr7_-_139876812 | 0.85 |
ENST00000397560.2
|
JHDM1D
|
lysine (K)-specific demethylase 7A |
| chr16_+_1031762 | 0.85 |
ENST00000293894.3
|
SOX8
|
SRY (sex determining region Y)-box 8 |
| chr12_+_56862301 | 0.83 |
ENST00000338146.5
|
SPRYD4
|
SPRY domain containing 4 |
| chr15_+_89181974 | 0.78 |
ENST00000306072.5
|
ISG20
|
interferon stimulated exonuclease gene 20kDa |
| chr13_+_52158610 | 0.76 |
ENST00000298125.5
|
WDFY2
|
WD repeat and FYVE domain containing 2 |
| chr16_+_3070356 | 0.76 |
ENST00000341627.5
ENST00000575124.1 ENST00000575836.1 |
TNFRSF12A
|
tumor necrosis factor receptor superfamily, member 12A |
| chr17_+_40440481 | 0.75 |
ENST00000590726.2
ENST00000452307.2 ENST00000444283.1 ENST00000588868.1 |
STAT5A
|
signal transducer and activator of transcription 5A |
| chr3_-_128840604 | 0.73 |
ENST00000476465.1
ENST00000315150.5 ENST00000393304.1 ENST00000393308.1 ENST00000393307.1 ENST00000393305.1 |
RAB43
|
RAB43, member RAS oncogene family |
| chr5_+_34656331 | 0.72 |
ENST00000265109.3
|
RAI14
|
retinoic acid induced 14 |
| chr8_+_26240666 | 0.72 |
ENST00000523949.1
|
BNIP3L
|
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chr5_+_34656529 | 0.72 |
ENST00000513974.1
ENST00000512629.1 |
RAI14
|
retinoic acid induced 14 |
| chr11_+_18416103 | 0.72 |
ENST00000543445.1
ENST00000430553.2 ENST00000396222.2 ENST00000535451.1 |
LDHA
|
lactate dehydrogenase A |
| chr2_+_201171372 | 0.71 |
ENST00000409140.3
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr14_-_62217779 | 0.68 |
ENST00000554254.1
|
HIF1A-AS2
|
HIF1A antisense RNA 2 |
| chr3_+_5020801 | 0.66 |
ENST00000256495.3
|
BHLHE40
|
basic helix-loop-helix family, member e40 |
| chr19_-_55895966 | 0.66 |
ENST00000444469.3
|
TMEM238
|
transmembrane protein 238 |
| chr9_+_19049372 | 0.66 |
ENST00000380527.1
|
RRAGA
|
Ras-related GTP binding A |
| chr11_+_69455855 | 0.66 |
ENST00000227507.2
ENST00000536559.1 |
CCND1
|
cyclin D1 |
| chr19_-_663147 | 0.66 |
ENST00000606702.1
|
RNF126
|
ring finger protein 126 |
| chr12_-_31478428 | 0.65 |
ENST00000543615.1
|
FAM60A
|
family with sequence similarity 60, member A |
| chr22_-_39548443 | 0.65 |
ENST00000401405.3
|
CBX7
|
chromobox homolog 7 |
| chr5_+_34656569 | 0.62 |
ENST00000428746.2
|
RAI14
|
retinoic acid induced 14 |
| chr11_+_18416133 | 0.62 |
ENST00000227157.4
ENST00000478970.2 ENST00000495052.1 |
LDHA
|
lactate dehydrogenase A |
| chr19_-_663171 | 0.62 |
ENST00000606896.1
ENST00000589762.2 |
RNF126
|
ring finger protein 126 |
| chr19_-_31840130 | 0.62 |
ENST00000558569.1
|
TSHZ3
|
teashirt zinc finger homeobox 3 |
| chr13_-_45151259 | 0.62 |
ENST00000493016.1
|
TSC22D1
|
TSC22 domain family, member 1 |
| chr10_+_112257596 | 0.61 |
ENST00000369583.3
|
DUSP5
|
dual specificity phosphatase 5 |
| chr10_-_126849626 | 0.61 |
ENST00000530884.1
|
CTBP2
|
C-terminal binding protein 2 |
| chr10_+_101419187 | 0.61 |
ENST00000370489.4
|
ENTPD7
|
ectonucleoside triphosphate diphosphohydrolase 7 |
| chr12_-_49730970 | 0.61 |
ENST00000334221.3
|
C1QL4
|
complement component 1, q subcomponent-like 4 |
| chr15_-_41408339 | 0.60 |
ENST00000401393.3
|
INO80
|
INO80 complex subunit |
| chr2_-_232328867 | 0.60 |
ENST00000453992.1
ENST00000417652.1 ENST00000454824.1 |
NCL
|
nucleolin |
| chr19_-_663277 | 0.59 |
ENST00000292363.5
|
RNF126
|
ring finger protein 126 |
| chr11_+_72929402 | 0.58 |
ENST00000393596.2
|
P2RY2
|
purinergic receptor P2Y, G-protein coupled, 2 |
| chr11_+_76494253 | 0.57 |
ENST00000333090.4
|
TSKU
|
tsukushi, small leucine rich proteoglycan |
| chr18_+_33877654 | 0.56 |
ENST00000257209.4
ENST00000445677.1 ENST00000590592.1 ENST00000359247.4 |
FHOD3
|
formin homology 2 domain containing 3 |
| chr19_-_1237990 | 0.56 |
ENST00000382477.2
ENST00000215376.6 ENST00000590083.1 |
C19orf26
|
chromosome 19 open reading frame 26 |
| chr11_-_6704513 | 0.56 |
ENST00000532203.1
ENST00000288937.6 |
MRPL17
|
mitochondrial ribosomal protein L17 |
| chr1_+_40420802 | 0.55 |
ENST00000372811.5
ENST00000420632.2 ENST00000434861.1 ENST00000372809.5 |
MFSD2A
|
major facilitator superfamily domain containing 2A |
| chr11_-_64646086 | 0.55 |
ENST00000320631.3
|
EHD1
|
EH-domain containing 1 |
| chr2_+_201171064 | 0.55 |
ENST00000451764.2
|
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr9_+_112542572 | 0.55 |
ENST00000374530.3
|
PALM2-AKAP2
|
PALM2-AKAP2 readthrough |
| chr1_+_44445643 | 0.55 |
ENST00000309519.7
|
B4GALT2
|
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr5_-_121413974 | 0.54 |
ENST00000231004.4
|
LOX
|
lysyl oxidase |
| chr7_-_92465868 | 0.54 |
ENST00000424848.2
|
CDK6
|
cyclin-dependent kinase 6 |
| chr1_+_209848749 | 0.54 |
ENST00000367029.4
|
G0S2
|
G0/G1switch 2 |
| chr5_+_34656450 | 0.54 |
ENST00000514527.1
|
RAI14
|
retinoic acid induced 14 |
| chr10_-_22292675 | 0.54 |
ENST00000376946.1
|
DNAJC1
|
DnaJ (Hsp40) homolog, subfamily C, member 1 |
| chr20_-_44540686 | 0.52 |
ENST00000477313.1
ENST00000542937.1 ENST00000372431.3 ENST00000354050.4 ENST00000420868.2 |
PLTP
|
phospholipid transfer protein |
| chr17_-_76713100 | 0.51 |
ENST00000585509.1
|
CYTH1
|
cytohesin 1 |
| chr5_-_131563501 | 0.51 |
ENST00000401867.1
ENST00000379086.1 ENST00000418055.1 ENST00000453286.1 ENST00000166534.4 |
P4HA2
|
prolyl 4-hydroxylase, alpha polypeptide II |
| chr2_+_177053307 | 0.51 |
ENST00000331462.4
|
HOXD1
|
homeobox D1 |
| chr19_+_1407733 | 0.50 |
ENST00000592453.1
|
DAZAP1
|
DAZ associated protein 1 |
| chr6_+_31865552 | 0.50 |
ENST00000469372.1
ENST00000497706.1 |
C2
|
complement component 2 |
| chr3_+_122785895 | 0.50 |
ENST00000316218.7
|
PDIA5
|
protein disulfide isomerase family A, member 5 |
| chr20_-_6103666 | 0.49 |
ENST00000536936.1
|
FERMT1
|
fermitin family member 1 |
| chr6_+_33257346 | 0.49 |
ENST00000374606.5
ENST00000374610.2 ENST00000374607.1 |
PFDN6
|
prefoldin subunit 6 |
| chr7_-_139876734 | 0.49 |
ENST00000006967.5
|
JHDM1D
|
lysine (K)-specific demethylase 7A |
| chr1_-_19229218 | 0.48 |
ENST00000432718.1
|
ALDH4A1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr22_+_24236191 | 0.48 |
ENST00000215754.7
|
MIF
|
macrophage migration inhibitory factor (glycosylation-inhibiting factor) |
| chr1_-_43638168 | 0.48 |
ENST00000431635.2
|
EBNA1BP2
|
EBNA1 binding protein 2 |
| chr7_-_130597935 | 0.48 |
ENST00000447307.1
ENST00000418546.1 |
MIR29B1
|
microRNA 29a |
| chr3_-_19988462 | 0.47 |
ENST00000344838.4
|
EFHB
|
EF-hand domain family, member B |
| chr2_+_28113583 | 0.47 |
ENST00000344773.2
ENST00000379624.1 ENST00000342045.2 ENST00000379632.2 ENST00000361704.2 |
BRE
|
brain and reproductive organ-expressed (TNFRSF1A modulator) |
| chr9_+_130159433 | 0.46 |
ENST00000451404.1
|
SLC2A8
|
solute carrier family 2 (facilitated glucose transporter), member 8 |
| chr8_-_37756972 | 0.46 |
ENST00000330843.4
ENST00000522727.1 ENST00000287263.4 |
RAB11FIP1
|
RAB11 family interacting protein 1 (class I) |
| chr11_-_59436453 | 0.46 |
ENST00000300146.9
|
PATL1
|
protein associated with topoisomerase II homolog 1 (yeast) |
| chr22_-_20255212 | 0.46 |
ENST00000416372.1
|
RTN4R
|
reticulon 4 receptor |
| chr20_+_20348740 | 0.46 |
ENST00000310227.1
|
INSM1
|
insulinoma-associated 1 |
| chr17_+_40985407 | 0.46 |
ENST00000586114.1
ENST00000590720.1 ENST00000585805.1 ENST00000541124.1 ENST00000441946.2 ENST00000591152.1 ENST00000589469.1 ENST00000293362.3 ENST00000592169.1 |
PSME3
|
proteasome (prosome, macropain) activator subunit 3 (PA28 gamma; Ki) |
| chr1_-_31712401 | 0.45 |
ENST00000373736.2
|
NKAIN1
|
Na+/K+ transporting ATPase interacting 1 |
| chr17_-_882966 | 0.45 |
ENST00000336868.3
|
NXN
|
nucleoredoxin |
| chr18_-_33077556 | 0.45 |
ENST00000589273.1
ENST00000586489.1 |
INO80C
|
INO80 complex subunit C |
| chr11_+_57435219 | 0.44 |
ENST00000527985.1
ENST00000287169.3 |
ZDHHC5
|
zinc finger, DHHC-type containing 5 |
| chr17_+_72428218 | 0.44 |
ENST00000392628.2
|
GPRC5C
|
G protein-coupled receptor, family C, group 5, member C |
| chr5_-_131563474 | 0.43 |
ENST00000417528.1
|
P4HA2
|
prolyl 4-hydroxylase, alpha polypeptide II |
| chr19_+_19516561 | 0.43 |
ENST00000457895.2
|
GATAD2A
|
GATA zinc finger domain containing 2A |
| chr2_+_111878483 | 0.43 |
ENST00000308659.8
ENST00000357757.2 ENST00000393253.2 ENST00000337565.5 ENST00000393256.3 |
BCL2L11
|
BCL2-like 11 (apoptosis facilitator) |
| chr11_-_124806297 | 0.43 |
ENST00000298251.4
|
HEPACAM
|
hepatic and glial cell adhesion molecule |
| chr2_+_201170999 | 0.43 |
ENST00000439395.1
ENST00000444012.1 |
SPATS2L
|
spermatogenesis associated, serine-rich 2-like |
| chr18_-_33077868 | 0.43 |
ENST00000590757.1
ENST00000592173.1 ENST00000441607.2 ENST00000587450.1 ENST00000589258.1 |
INO80C
RP11-322E11.6
|
INO80 complex subunit C Uncharacterized protein |
| chr6_+_33257427 | 0.42 |
ENST00000463584.1
|
PFDN6
|
prefoldin subunit 6 |
| chr11_-_64511789 | 0.42 |
ENST00000419843.1
ENST00000394430.1 |
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr14_-_88459182 | 0.42 |
ENST00000544807.2
|
GALC
|
galactosylceramidase |
| chr8_+_126442563 | 0.42 |
ENST00000311922.3
|
TRIB1
|
tribbles pseudokinase 1 |
| chr22_+_20104947 | 0.42 |
ENST00000402752.1
|
RANBP1
|
RAN binding protein 1 |
| chr1_-_8938736 | 0.42 |
ENST00000234590.4
|
ENO1
|
enolase 1, (alpha) |
| chr9_+_112542591 | 0.42 |
ENST00000483909.1
ENST00000314527.4 ENST00000413420.1 ENST00000302798.7 ENST00000555236.1 ENST00000510514.5 |
PALM2
PALM2-AKAP2
AKAP2
|
paralemmin 2 PALM2-AKAP2 readthrough A kinase (PRKA) anchor protein 2 |
| chr6_+_151561085 | 0.42 |
ENST00000402676.2
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr1_-_43637915 | 0.42 |
ENST00000236051.2
|
EBNA1BP2
|
EBNA1 binding protein 2 |
| chr17_+_46048471 | 0.42 |
ENST00000578018.1
ENST00000579175.1 |
CDK5RAP3
|
CDK5 regulatory subunit associated protein 3 |
| chr2_+_113403434 | 0.42 |
ENST00000272542.3
|
SLC20A1
|
solute carrier family 20 (phosphate transporter), member 1 |
| chr12_+_112451222 | 0.41 |
ENST00000552052.1
|
ERP29
|
endoplasmic reticulum protein 29 |
| chr2_+_10183651 | 0.41 |
ENST00000305883.1
|
KLF11
|
Kruppel-like factor 11 |
| chr19_+_55987998 | 0.41 |
ENST00000591164.1
|
ZNF628
|
zinc finger protein 628 |
| chr12_-_51422017 | 0.41 |
ENST00000394904.3
|
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr6_-_33256664 | 0.41 |
ENST00000444176.1
|
WDR46
|
WD repeat domain 46 |
| chr6_+_151561506 | 0.40 |
ENST00000253332.1
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr12_+_15475462 | 0.40 |
ENST00000543886.1
ENST00000348962.2 |
PTPRO
|
protein tyrosine phosphatase, receptor type, O |
| chr1_+_113217073 | 0.40 |
ENST00000369645.1
|
MOV10
|
Mov10, Moloney leukemia virus 10, homolog (mouse) |
| chr17_-_2615031 | 0.40 |
ENST00000576885.1
ENST00000574426.2 |
CLUH
|
clustered mitochondria (cluA/CLU1) homolog |
| chr5_+_74807886 | 0.40 |
ENST00000514296.1
|
POLK
|
polymerase (DNA directed) kappa |
| chr21_+_26934165 | 0.40 |
ENST00000456917.1
|
MIR155HG
|
MIR155 host gene (non-protein coding) |
| chr14_-_88459503 | 0.39 |
ENST00000393568.4
ENST00000261304.2 |
GALC
|
galactosylceramidase |
| chr22_+_40390930 | 0.39 |
ENST00000333407.6
|
FAM83F
|
family with sequence similarity 83, member F |
| chr19_-_17799135 | 0.39 |
ENST00000552293.1
ENST00000551649.1 ENST00000550896.1 |
UNC13A
|
unc-13 homolog A (C. elegans) |
| chrX_+_24483338 | 0.39 |
ENST00000379162.4
ENST00000441463.2 |
PDK3
|
pyruvate dehydrogenase kinase, isozyme 3 |
| chr1_+_1167594 | 0.38 |
ENST00000379198.2
|
B3GALT6
|
UDP-Gal:betaGal beta 1,3-galactosyltransferase polypeptide 6 |
| chr19_+_18942720 | 0.38 |
ENST00000262803.5
|
UPF1
|
UPF1 regulator of nonsense transcripts homolog (yeast) |
| chr6_+_42896865 | 0.38 |
ENST00000372836.4
ENST00000394142.3 |
CNPY3
|
canopy FGF signaling regulator 3 |
| chr3_+_183415558 | 0.38 |
ENST00000305135.5
|
YEATS2
|
YEATS domain containing 2 |
| chr3_-_183543301 | 0.38 |
ENST00000318631.3
ENST00000431348.1 |
MAP6D1
|
MAP6 domain containing 1 |
| chr17_+_46048497 | 0.37 |
ENST00000583352.1
|
CDK5RAP3
|
CDK5 regulatory subunit associated protein 3 |
| chr17_-_36831156 | 0.37 |
ENST00000325814.5
|
C17orf96
|
chromosome 17 open reading frame 96 |
| chr11_+_107461948 | 0.37 |
ENST00000265840.7
ENST00000443271.2 |
ELMOD1
|
ELMO/CED-12 domain containing 1 |
| chr18_-_33077942 | 0.37 |
ENST00000334598.7
|
INO80C
|
INO80 complex subunit C |
| chr19_+_5720666 | 0.37 |
ENST00000381624.3
ENST00000381614.2 |
CATSPERD
|
catsper channel auxiliary subunit delta |
| chr8_-_124553437 | 0.36 |
ENST00000517956.1
ENST00000443022.2 |
FBXO32
|
F-box protein 32 |
| chr10_-_16859442 | 0.36 |
ENST00000602389.1
ENST00000345264.5 |
RSU1
|
Ras suppressor protein 1 |
| chr16_-_1031520 | 0.36 |
ENST00000568394.1
ENST00000565467.1 |
RP11-161M6.2
|
RP11-161M6.2 |
| chr10_+_99258625 | 0.36 |
ENST00000370664.3
|
UBTD1
|
ubiquitin domain containing 1 |
| chr11_-_67771513 | 0.36 |
ENST00000227471.2
|
UNC93B1
|
unc-93 homolog B1 (C. elegans) |
| chr19_-_44258770 | 0.36 |
ENST00000601925.1
ENST00000602222.1 ENST00000599804.1 |
SMG9
|
SMG9 nonsense mediated mRNA decay factor |
| chr1_-_201476274 | 0.36 |
ENST00000340006.2
|
CSRP1
|
cysteine and glycine-rich protein 1 |
| chr17_+_46048376 | 0.36 |
ENST00000338399.4
|
CDK5RAP3
|
CDK5 regulatory subunit associated protein 3 |
| chrX_+_152912616 | 0.35 |
ENST00000342782.3
|
DUSP9
|
dual specificity phosphatase 9 |
| chr19_-_47354023 | 0.35 |
ENST00000601649.1
ENST00000599990.1 ENST00000352203.4 |
AP2S1
|
adaptor-related protein complex 2, sigma 1 subunit |
| chr22_-_30987837 | 0.35 |
ENST00000335214.6
|
PES1
|
pescadillo ribosomal biogenesis factor 1 |
| chrX_-_53254358 | 0.35 |
ENST00000404049.3
|
KDM5C
|
lysine (K)-specific demethylase 5C |
| chr7_+_116312411 | 0.35 |
ENST00000456159.1
ENST00000397752.3 ENST00000318493.6 |
MET
|
met proto-oncogene |
| chr17_+_56833184 | 0.34 |
ENST00000308249.2
|
PPM1E
|
protein phosphatase, Mg2+/Mn2+ dependent, 1E |
| chr1_+_230203010 | 0.34 |
ENST00000541865.1
|
GALNT2
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-T2) |
| chr3_+_32433363 | 0.34 |
ENST00000465248.1
|
CMTM7
|
CKLF-like MARVEL transmembrane domain containing 7 |
| chr19_+_2977444 | 0.34 |
ENST00000246112.4
ENST00000453329.1 ENST00000482627.1 ENST00000452088.1 |
TLE6
|
transducin-like enhancer of split 6 (E(sp1) homolog, Drosophila) |
| chr11_-_64511575 | 0.34 |
ENST00000431822.1
ENST00000377486.3 ENST00000394432.3 |
RASGRP2
|
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
| chr2_-_232329186 | 0.34 |
ENST00000322723.4
|
NCL
|
nucleolin |
| chr20_+_56964169 | 0.34 |
ENST00000475243.1
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr9_+_130159504 | 0.34 |
ENST00000373352.1
ENST00000373360.3 |
SLC2A8
|
solute carrier family 2 (facilitated glucose transporter), member 8 |
| chr11_-_3013482 | 0.34 |
ENST00000529361.1
ENST00000528968.1 ENST00000534372.1 ENST00000531291.1 ENST00000526842.1 |
NAP1L4
|
nucleosome assembly protein 1-like 4 |
| chr3_+_100211412 | 0.33 |
ENST00000323523.4
ENST00000403410.1 ENST00000449609.1 |
TMEM45A
|
transmembrane protein 45A |
| chr12_+_7023735 | 0.33 |
ENST00000538763.1
ENST00000544774.1 ENST00000545045.2 |
ENO2
|
enolase 2 (gamma, neuronal) |
| chr10_-_16859361 | 0.33 |
ENST00000377921.3
|
RSU1
|
Ras suppressor protein 1 |
| chr15_-_30261066 | 0.33 |
ENST00000558447.1
|
TJP1
|
tight junction protein 1 |
| chr1_+_154193325 | 0.33 |
ENST00000428931.1
ENST00000441890.1 ENST00000271877.7 ENST00000412596.1 ENST00000368504.1 ENST00000437652.1 |
UBAP2L
|
ubiquitin associated protein 2-like |
| chr5_-_107006596 | 0.33 |
ENST00000333274.6
|
EFNA5
|
ephrin-A5 |
| chr17_+_57642886 | 0.33 |
ENST00000251241.4
ENST00000451169.2 ENST00000425628.3 ENST00000584385.1 ENST00000580030.1 |
DHX40
|
DEAH (Asp-Glu-Ala-His) box polypeptide 40 |
| chr19_-_10530784 | 0.32 |
ENST00000593124.1
|
CDC37
|
cell division cycle 37 |
| chr22_-_30987849 | 0.32 |
ENST00000402284.3
ENST00000354694.7 |
PES1
|
pescadillo ribosomal biogenesis factor 1 |
| chr19_-_17622684 | 0.32 |
ENST00000596643.1
|
CTD-3131K8.2
|
CTD-3131K8.2 |
| chr19_+_18942761 | 0.32 |
ENST00000599848.1
|
UPF1
|
UPF1 regulator of nonsense transcripts homolog (yeast) |
| chr19_+_50180409 | 0.32 |
ENST00000391851.4
|
PRMT1
|
protein arginine methyltransferase 1 |
| chr14_-_20929624 | 0.32 |
ENST00000398020.4
ENST00000250489.4 |
TMEM55B
|
transmembrane protein 55B |
| chr17_-_42277203 | 0.32 |
ENST00000587097.1
|
ATXN7L3
|
ataxin 7-like 3 |
| chr3_-_127309550 | 0.32 |
ENST00000296210.7
ENST00000355552.3 |
TPRA1
|
transmembrane protein, adipocyte asscociated 1 |
| chrX_-_48814810 | 0.32 |
ENST00000376488.3
ENST00000396743.3 ENST00000156084.4 |
OTUD5
|
OTU domain containing 5 |
| chr17_-_37557846 | 0.32 |
ENST00000394294.3
ENST00000583610.1 ENST00000264658.6 |
FBXL20
|
F-box and leucine-rich repeat protein 20 |
| chr4_-_103266355 | 0.32 |
ENST00000424970.2
|
SLC39A8
|
solute carrier family 39 (zinc transporter), member 8 |
| chr12_+_6977258 | 0.32 |
ENST00000488464.2
ENST00000535434.1 ENST00000493987.1 |
TPI1
|
triosephosphate isomerase 1 |
| chr3_-_185826718 | 0.32 |
ENST00000413301.1
ENST00000421809.1 |
ETV5
|
ets variant 5 |
| chr14_+_65453432 | 0.32 |
ENST00000246166.2
|
FNTB
|
farnesyltransferase, CAAX box, beta |
| chr8_+_95908041 | 0.32 |
ENST00000396113.1
ENST00000519136.1 |
NDUFAF6
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr2_+_27665289 | 0.32 |
ENST00000407293.1
|
KRTCAP3
|
keratinocyte associated protein 3 |
| chr19_-_47354082 | 0.32 |
ENST00000593442.1
ENST00000263270.6 |
AP2S1
|
adaptor-related protein complex 2, sigma 1 subunit |
| chr9_+_130159471 | 0.32 |
ENST00000419917.1
|
SLC2A8
|
solute carrier family 2 (facilitated glucose transporter), member 8 |
| chr6_+_34204642 | 0.31 |
ENST00000347617.6
ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1
|
high mobility group AT-hook 1 |
| chr17_-_58603482 | 0.31 |
ENST00000585368.1
|
APPBP2
|
amyloid beta precursor protein (cytoplasmic tail) binding protein 2 |
| chr7_+_75677354 | 0.30 |
ENST00000461263.2
ENST00000315758.5 ENST00000443006.1 |
MDH2
|
malate dehydrogenase 2, NAD (mitochondrial) |
| chr7_+_6048856 | 0.30 |
ENST00000223029.3
ENST00000400479.2 ENST00000395236.2 |
AIMP2
|
aminoacyl tRNA synthetase complex-interacting multifunctional protein 2 |
| chr12_+_7023491 | 0.30 |
ENST00000541477.1
ENST00000229277.1 |
ENO2
|
enolase 2 (gamma, neuronal) |
| chr3_+_47021168 | 0.30 |
ENST00000450053.3
ENST00000292309.5 ENST00000383740.2 |
NBEAL2
|
neurobeachin-like 2 |
| chr18_+_43913919 | 0.30 |
ENST00000587853.1
|
RNF165
|
ring finger protein 165 |
| chr3_+_133292759 | 0.30 |
ENST00000431519.2
|
CDV3
|
CDV3 homolog (mouse) |
| chr1_+_87170577 | 0.30 |
ENST00000482504.1
|
SH3GLB1
|
SH3-domain GRB2-like endophilin B1 |
| chr1_-_201476220 | 0.30 |
ENST00000526723.1
ENST00000524951.1 |
CSRP1
|
cysteine and glycine-rich protein 1 |
| chr17_+_80416050 | 0.30 |
ENST00000579198.1
ENST00000390006.4 ENST00000580296.1 |
NARF
|
nuclear prelamin A recognition factor |
| chr7_+_75677465 | 0.29 |
ENST00000432020.2
|
MDH2
|
malate dehydrogenase 2, NAD (mitochondrial) |
| chr19_-_39523165 | 0.29 |
ENST00000509137.2
ENST00000292853.4 |
FBXO27
|
F-box protein 27 |
| chr3_-_185826855 | 0.29 |
ENST00000306376.5
|
ETV5
|
ets variant 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ARNT
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0071629 | cytoplasm-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071629) |
| 0.6 | 1.7 | GO:2000564 | CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.5 | 2.7 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.4 | 1.2 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.3 | 1.0 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.3 | 0.8 | GO:0072034 | renal vesicle induction(GO:0072034) |
| 0.3 | 1.9 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.2 | 0.6 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.2 | 2.6 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.2 | 1.0 | GO:0035802 | adrenal cortex development(GO:0035801) adrenal cortex formation(GO:0035802) |
| 0.2 | 0.6 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.2 | 0.6 | GO:0070407 | oxidation-dependent protein catabolic process(GO:0070407) |
| 0.2 | 0.7 | GO:0000294 | nuclear-transcribed mRNA catabolic process, endonucleolytic cleavage-dependent decay(GO:0000294) |
| 0.2 | 0.2 | GO:2000785 | regulation of vacuole organization(GO:0044088) regulation of autophagosome assembly(GO:2000785) |
| 0.2 | 0.8 | GO:0048133 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.2 | 0.7 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.2 | 1.6 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.2 | 0.9 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.2 | 1.2 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.2 | 0.8 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.1 | 0.4 | GO:0060139 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.1 | 0.4 | GO:0035572 | N-terminal peptidyl-alanine methylation(GO:0018011) N-terminal peptidyl-alanine trimethylation(GO:0018012) N-terminal peptidyl-glycine methylation(GO:0018013) N-terminal peptidyl-proline dimethylation(GO:0018016) peptidyl-alanine modification(GO:0018194) N-terminal peptidyl-proline methylation(GO:0035568) N-terminal peptidyl-serine methylation(GO:0035570) N-terminal peptidyl-serine dimethylation(GO:0035572) N-terminal peptidyl-serine trimethylation(GO:0035573) |
| 0.1 | 0.4 | GO:0045659 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.1 | 0.4 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) |
| 0.1 | 2.7 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.1 | 0.5 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.1 | 1.1 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.1 | 0.4 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.1 | 0.5 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.1 | 0.3 | GO:0043095 | regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 0.1 | 0.3 | GO:1903630 | regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 0.1 | 0.8 | GO:0035694 | mitochondrial protein catabolic process(GO:0035694) |
| 0.1 | 0.5 | GO:0048817 | negative regulation of hair follicle maturation(GO:0048817) regulation of transforming growth factor-beta secretion(GO:2001201) |
| 0.1 | 1.5 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.1 | 0.4 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 0.1 | 0.6 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.1 | 0.6 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.1 | 0.3 | GO:0034164 | negative regulation of toll-like receptor 9 signaling pathway(GO:0034164) |
| 0.1 | 0.6 | GO:0046985 | positive regulation of hemoglobin biosynthetic process(GO:0046985) |
| 0.1 | 0.5 | GO:0003357 | noradrenergic neuron differentiation(GO:0003357) |
| 0.1 | 0.1 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.1 | 0.4 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.1 | 0.3 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.1 | 0.2 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.1 | 0.8 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 | 0.3 | GO:1903778 | protein localization to vacuolar membrane(GO:1903778) |
| 0.1 | 0.8 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.1 | 0.2 | GO:0021722 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.1 | 0.6 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.1 | 1.3 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 0.5 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.1 | 0.5 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.1 | 0.4 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.1 | 0.4 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.1 | 0.7 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.1 | 0.7 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.1 | 0.1 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.1 | 0.3 | GO:0086021 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) |
| 0.1 | 0.3 | GO:0018277 | protein deamination(GO:0018277) |
| 0.1 | 0.8 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.1 | 0.3 | GO:0035986 | senescence-associated heterochromatin focus assembly(GO:0035986) oncogene-induced cell senescence(GO:0090402) |
| 0.1 | 1.4 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.1 | 0.2 | GO:0046166 | glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.1 | 0.4 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.1 | 0.9 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.1 | 0.2 | GO:0016333 | morphogenesis of follicular epithelium(GO:0016333) establishment or maintenance of polarity of follicular epithelium(GO:0016334) establishment of planar polarity of follicular epithelium(GO:0042247) |
| 0.1 | 0.5 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.1 | 0.7 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.1 | 0.5 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) |
| 0.1 | 0.5 | GO:0045656 | negative regulation of monocyte differentiation(GO:0045656) |
| 0.1 | 0.2 | GO:0000472 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.1 | 0.3 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.1 | 0.3 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 0.2 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.1 | 0.2 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) 7-methylguanosine RNA capping(GO:0009452) RNA capping(GO:0036260) |
| 0.1 | 0.4 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.0 | 0.3 | GO:2000259 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.0 | 0.1 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.2 | GO:0014859 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.0 | 0.6 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 0.1 | GO:0038162 | erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.0 | 0.2 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.0 | 0.1 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.3 | GO:0072619 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.6 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.7 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.7 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.3 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.2 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.2 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.1 | GO:1902512 | positive regulation of apoptotic DNA fragmentation(GO:1902512) |
| 0.0 | 0.9 | GO:0019511 | peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.2 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.6 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.5 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.2 | GO:0046452 | dihydrofolate metabolic process(GO:0046452) |
| 0.0 | 0.5 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.3 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 1.1 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 | 0.3 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 | 0.8 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.4 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 1.1 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.0 | 0.5 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.7 | GO:0010603 | regulation of cytoplasmic mRNA processing body assembly(GO:0010603) |
| 0.0 | 0.3 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.2 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.0 | 0.2 | GO:0048073 | regulation of eye pigmentation(GO:0048073) |
| 0.0 | 0.1 | GO:0021784 | postganglionic parasympathetic fiber development(GO:0021784) |
| 0.0 | 0.3 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.1 | GO:0034402 | recruitment of 3'-end processing factors to RNA polymerase II holoenzyme complex(GO:0034402) |
| 0.0 | 0.2 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.6 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 | 0.2 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.1 | GO:1902824 | positive regulation of late endosome to lysosome transport(GO:1902824) |
| 0.0 | 0.8 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.4 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.1 | GO:0044417 | translocation of peptides or proteins into host(GO:0042000) translocation of peptides or proteins into host cell cytoplasm(GO:0044053) translocation of molecules into host(GO:0044417) translocation of peptides or proteins into other organism involved in symbiotic interaction(GO:0051808) translocation of molecules into other organism involved in symbiotic interaction(GO:0051836) |
| 0.0 | 0.1 | GO:0006424 | glutamyl-tRNA aminoacylation(GO:0006424) |
| 0.0 | 0.2 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 | 0.2 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.0 | 0.2 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.0 | 0.2 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.0 | 0.5 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.0 | 0.2 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 1.1 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.2 | GO:0097168 | mesenchymal stem cell proliferation(GO:0097168) regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.0 | 0.3 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.3 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.3 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.3 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.2 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.0 | 0.5 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.2 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.0 | 0.6 | GO:0060856 | establishment of blood-brain barrier(GO:0060856) |
| 0.0 | 0.8 | GO:0032802 | low-density lipoprotein particle receptor catabolic process(GO:0032802) |
| 0.0 | 0.2 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.4 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.1 | GO:1904100 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.0 | 0.6 | GO:0034656 | nucleobase-containing small molecule catabolic process(GO:0034656) |
| 0.0 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.2 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.3 | GO:0060019 | radial glial cell differentiation(GO:0060019) |
| 0.0 | 0.2 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 0.2 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.3 | GO:0010820 | positive regulation of T cell chemotaxis(GO:0010820) |
| 0.0 | 0.1 | GO:0072287 | metanephric distal tubule morphogenesis(GO:0072287) |
| 0.0 | 0.2 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.0 | 0.5 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.1 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.0 | 0.7 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.0 | 0.2 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.2 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.0 | 0.1 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.0 | 0.4 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.3 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.3 | GO:1904322 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 0.1 | GO:1902430 | negative regulation of beta-amyloid formation(GO:1902430) |
| 0.0 | 0.2 | GO:0046654 | tetrahydrofolate biosynthetic process(GO:0046654) |
| 0.0 | 1.7 | GO:0001658 | branching involved in ureteric bud morphogenesis(GO:0001658) |
| 0.0 | 0.4 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.6 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.0 | 0.0 | GO:0044533 | induction of programmed cell death(GO:0012502) positive regulation of apoptotic process in other organism(GO:0044533) positive regulation by symbiont of host programmed cell death(GO:0052042) positive regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052330) positive regulation by organism of apoptotic process in other organism involved in symbiotic interaction(GO:0052501) |
| 0.0 | 0.1 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.0 | 0.1 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.0 | 0.5 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.5 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.5 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 0.1 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.2 | GO:0051963 | regulation of synapse assembly(GO:0051963) positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.1 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.0 | 0.6 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
| 0.0 | 0.2 | GO:0051611 | negative regulation of neurotransmitter uptake(GO:0051581) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.3 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.0 | 0.3 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.1 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.0 | 0.2 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.0 | 0.1 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.0 | 0.1 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 0.1 | GO:0048842 | positive regulation of axon extension involved in axon guidance(GO:0048842) regulation of endothelial cell chemotaxis(GO:2001026) positive regulation of endothelial cell chemotaxis(GO:2001028) |
| 0.0 | 0.1 | GO:0070272 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 | 0.1 | GO:0031126 | snoRNA 3'-end processing(GO:0031126) |
| 0.0 | 0.2 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.0 | 0.2 | GO:0008635 | activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c(GO:0008635) |
| 0.0 | 0.2 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.4 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.1 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.9 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.2 | GO:0034472 | snRNA 3'-end processing(GO:0034472) |
| 0.0 | 0.3 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.1 | GO:1904327 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.0 | 0.1 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.0 | 0.4 | GO:0017145 | stem cell division(GO:0017145) |
| 0.0 | 0.1 | GO:0044800 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 | 0.1 | GO:0000479 | endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.0 | 0.1 | GO:0015887 | biotin transport(GO:0015878) pantothenate transmembrane transport(GO:0015887) |
| 0.0 | 0.5 | GO:0060384 | innervation(GO:0060384) |
| 0.0 | 0.5 | GO:0002209 | behavioral fear response(GO:0001662) behavioral defense response(GO:0002209) |
| 0.0 | 0.3 | GO:0030220 | platelet formation(GO:0030220) |
| 0.0 | 0.2 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.0 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.0 | 0.0 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.2 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.1 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.1 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.0 | 0.0 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.0 | 0.2 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.0 | 0.3 | GO:0060338 | regulation of type I interferon-mediated signaling pathway(GO:0060338) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.3 | GO:0016578 | histone deubiquitination(GO:0016578) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.2 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.3 | 2.6 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.8 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 0.7 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.1 | 0.9 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.1 | 0.6 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.1 | 0.5 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.1 | 0.7 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.1 | 0.3 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.1 | 0.4 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.1 | 0.5 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 0.4 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 0.3 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.1 | 1.8 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.1 | 0.2 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 0.2 | GO:0030689 | Noc complex(GO:0030689) |
| 0.1 | 0.5 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 0.7 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.2 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.4 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.2 | GO:0042643 | actomyosin, actin portion(GO:0042643) |
| 0.0 | 0.4 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.2 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.9 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.0 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.0 | 0.4 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 1.1 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.3 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.4 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.2 | GO:0000333 | telomerase catalytic core complex(GO:0000333) |
| 0.0 | 0.5 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.9 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.0 | 0.8 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.4 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 2.7 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.2 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.6 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.6 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.2 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.1 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.8 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.2 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.2 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.8 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.3 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.2 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.8 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.1 | GO:0002133 | polycystin complex(GO:0002133) |
| 0.0 | 0.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.6 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.3 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.1 | GO:0031984 | organelle subcompartment(GO:0031984) |
| 0.0 | 0.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.1 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 1.2 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.2 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.3 | GO:0098563 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.3 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.6 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.2 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 0.2 | GO:0097025 | lateral loop(GO:0043219) MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.5 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.0 | 0.4 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 1.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.1 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.9 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:0004618 | phosphoglycerate kinase activity(GO:0004618) |
| 0.9 | 2.7 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.3 | 2.6 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.2 | 1.2 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.2 | 0.6 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.2 | 0.8 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.2 | 0.6 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.2 | 0.6 | GO:0070364 | mitochondrial light strand promoter anti-sense binding(GO:0070361) mitochondrial heavy strand promoter anti-sense binding(GO:0070362) mitochondrial heavy strand promoter sense binding(GO:0070364) |
| 0.2 | 0.5 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.2 | 0.5 | GO:0038131 | neuregulin receptor activity(GO:0038131) |
| 0.1 | 0.4 | GO:0097158 | pre-mRNA intronic pyrimidine-rich binding(GO:0097158) |
| 0.1 | 0.6 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 1.4 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.5 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.1 | 0.9 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.1 | 0.4 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.1 | 0.3 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 0.1 | 0.9 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.1 | 0.3 | GO:0044549 | GTP cyclohydrolase binding(GO:0044549) |
| 0.1 | 1.6 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.1 | 0.1 | GO:0004818 | glutamate-tRNA ligase activity(GO:0004818) |
| 0.1 | 0.5 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.1 | 0.2 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) |
| 0.1 | 0.7 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.1 | 0.5 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.1 | 0.3 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.1 | 0.2 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.1 | 0.7 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 0.6 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.1 | 0.2 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.1 | 0.2 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 0.1 | 1.0 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.1 | 0.4 | GO:0015086 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.1 | 0.2 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.1 | 1.5 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 0.6 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.1 | 0.5 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.1 | 0.2 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.1 | 0.3 | GO:0086020 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) |
| 0.1 | 0.5 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.6 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.1 | 1.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.5 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.3 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.2 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.4 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.1 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.8 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.5 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.5 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.1 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.0 | 0.2 | GO:0004637 | phosphoribosylamine-glycine ligase activity(GO:0004637) phosphoribosylformylglycinamidine cyclo-ligase activity(GO:0004641) phosphoribosylglycinamide formyltransferase activity(GO:0004644) |
| 0.0 | 0.0 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.2 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.1 | GO:0009383 | rRNA (cytosine-C5-)-methyltransferase activity(GO:0009383) |
| 0.0 | 0.3 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.1 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 1.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.5 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.3 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.0 | 0.1 | GO:0000035 | acyl binding(GO:0000035) phosphopantetheine binding(GO:0031177) |
| 0.0 | 0.3 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 2.1 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.7 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.9 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.3 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.6 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.2 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 1.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0003968 | RNA-directed RNA polymerase activity(GO:0003968) |
| 0.0 | 1.2 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.2 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.2 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 1.1 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.3 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 3.0 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.2 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.0 | 0.6 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.3 | GO:0032407 | MutSalpha complex binding(GO:0032407) |
| 0.0 | 1.0 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.3 | GO:0016273 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.3 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.4 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.2 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.1 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.0 | 0.1 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) cupric ion binding(GO:1903135) |
| 0.0 | 0.2 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.4 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 0.4 | GO:0055106 | ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.2 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.1 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 0.2 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.4 | GO:0035325 | Toll-like receptor binding(GO:0035325) |
| 0.0 | 0.6 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.2 | GO:0001016 | RNA polymerase III regulatory region DNA binding(GO:0001016) |
| 0.0 | 0.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.3 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.2 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.5 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 1.5 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 0.2 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.1 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.7 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.3 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.2 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.0 | 0.6 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.2 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.0 | 0.1 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.7 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.3 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.4 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.2 | GO:0005351 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) |
| 0.0 | 0.3 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.0 | 0.3 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.0 | GO:0032551 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.1 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.0 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.4 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.3 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0015295 | solute:proton symporter activity(GO:0015295) |
| 0.0 | 0.2 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.5 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.2 | GO:0000150 | recombinase activity(GO:0000150) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.9 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 2.2 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.8 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 2.6 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.6 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.8 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.4 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.1 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.4 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.4 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 1.3 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.9 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.1 | 5.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 1.5 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 0.7 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.1 | 1.0 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 1.9 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.1 | 1.7 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 1.1 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.9 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 1.2 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 2.7 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.8 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.8 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 1.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.5 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.3 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.5 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.5 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.6 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.5 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.3 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.7 | REACTOME PREFOLDIN MEDIATED TRANSFER OF SUBSTRATE TO CCT TRIC | Genes involved in Prefoldin mediated transfer of substrate to CCT/TriC |
| 0.0 | 0.7 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.2 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.3 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.4 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.8 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.2 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.5 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.2 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |