Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
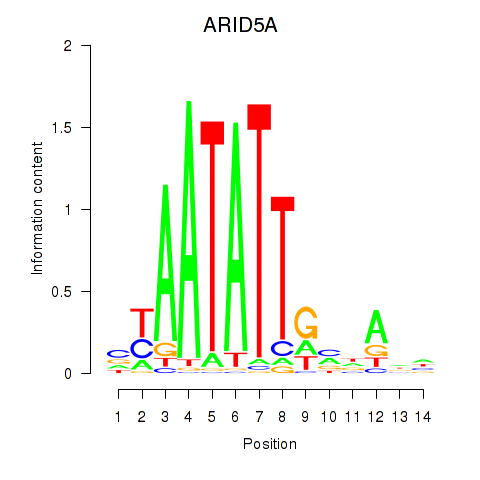
Results for ARID5A
Z-value: 0.87
Transcription factors associated with ARID5A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ARID5A
|
ENSG00000196843.11 | AT-rich interaction domain 5A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ARID5A | hg19_v2_chr2_+_97202480_97202499 | 0.50 | 5.0e-01 | Click! |
Activity profile of ARID5A motif
Sorted Z-values of ARID5A motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_79115503 | 0.96 |
ENST00000370747.4
ENST00000438486.1 ENST00000545124.1 |
IFI44
|
interferon-induced protein 44 |
| chr7_-_92777606 | 0.90 |
ENST00000437805.1
ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L
|
sterile alpha motif domain containing 9-like |
| chrX_+_41583408 | 0.65 |
ENST00000302548.4
|
GPR82
|
G protein-coupled receptor 82 |
| chr1_+_948803 | 0.58 |
ENST00000379389.4
|
ISG15
|
ISG15 ubiquitin-like modifier |
| chr1_-_89531041 | 0.55 |
ENST00000370473.4
|
GBP1
|
guanylate binding protein 1, interferon-inducible |
| chr18_-_61329118 | 0.46 |
ENST00000332821.8
ENST00000283752.5 |
SERPINB3
|
serpin peptidase inhibitor, clade B (ovalbumin), member 3 |
| chr12_+_28605426 | 0.44 |
ENST00000542801.1
|
CCDC91
|
coiled-coil domain containing 91 |
| chr14_-_67878917 | 0.42 |
ENST00000216446.4
|
PLEK2
|
pleckstrin 2 |
| chr17_+_68047418 | 0.42 |
ENST00000586373.1
ENST00000588782.1 |
LINC01028
|
long intergenic non-protein coding RNA 1028 |
| chr10_+_106113515 | 0.40 |
ENST00000369704.3
ENST00000312902.5 |
CCDC147
|
coiled-coil domain containing 147 |
| chr1_-_89736434 | 0.40 |
ENST00000370459.3
|
GBP5
|
guanylate binding protein 5 |
| chr10_+_24755416 | 0.40 |
ENST00000396446.1
ENST00000396445.1 ENST00000376451.2 |
KIAA1217
|
KIAA1217 |
| chr6_+_32812568 | 0.39 |
ENST00000414474.1
|
PSMB9
|
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr12_-_96389702 | 0.38 |
ENST00000552509.1
|
HAL
|
histidine ammonia-lyase |
| chr19_-_44174330 | 0.36 |
ENST00000340093.3
|
PLAUR
|
plasminogen activator, urokinase receptor |
| chr1_-_89591749 | 0.35 |
ENST00000370466.3
|
GBP2
|
guanylate binding protein 2, interferon-inducible |
| chrX_+_135252050 | 0.34 |
ENST00000449474.1
ENST00000345434.3 |
FHL1
|
four and a half LIM domains 1 |
| chr4_+_69962212 | 0.33 |
ENST00000508661.1
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr6_+_30035307 | 0.33 |
ENST00000376765.2
ENST00000376763.1 |
PPP1R11
|
protein phosphatase 1, regulatory (inhibitor) subunit 11 |
| chr15_-_78913628 | 0.33 |
ENST00000348639.3
|
CHRNA3
|
cholinergic receptor, nicotinic, alpha 3 (neuronal) |
| chr5_+_61874696 | 0.31 |
ENST00000491184.2
|
LRRC70
|
leucine rich repeat containing 70 |
| chr19_-_48753028 | 0.31 |
ENST00000522431.1
|
CARD8
|
caspase recruitment domain family, member 8 |
| chr19_-_44174305 | 0.30 |
ENST00000601723.1
ENST00000339082.3 |
PLAUR
|
plasminogen activator, urokinase receptor |
| chrM_+_8366 | 0.30 |
ENST00000361851.1
|
MT-ATP8
|
mitochondrially encoded ATP synthase 8 |
| chr1_-_154600421 | 0.29 |
ENST00000368471.3
ENST00000292205.5 |
ADAR
|
adenosine deaminase, RNA-specific |
| chr12_+_75874984 | 0.28 |
ENST00000550491.1
|
GLIPR1
|
GLI pathogenesis-related 1 |
| chr19_+_30156400 | 0.27 |
ENST00000588833.1
|
PLEKHF1
|
pleckstrin homology domain containing, family F (with FYVE domain) member 1 |
| chr3_-_69171708 | 0.27 |
ENST00000420581.2
|
LMOD3
|
leiomodin 3 (fetal) |
| chr16_-_3422283 | 0.26 |
ENST00000399974.3
|
MTRNR2L4
|
MT-RNR2-like 4 |
| chr13_-_52703187 | 0.26 |
ENST00000355568.4
|
NEK5
|
NIMA-related kinase 5 |
| chrX_+_36246735 | 0.26 |
ENST00000378653.3
|
CXorf30
|
chromosome X open reading frame 30 |
| chr12_+_10460417 | 0.25 |
ENST00000381908.3
ENST00000336164.4 ENST00000350274.5 |
KLRD1
|
killer cell lectin-like receptor subfamily D, member 1 |
| chr2_+_149447783 | 0.25 |
ENST00000449013.1
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr12_-_10588539 | 0.25 |
ENST00000381902.2
ENST00000381901.1 ENST00000539033.1 |
KLRC2
NKG2-E
|
killer cell lectin-like receptor subfamily C, member 2 Uncharacterized protein |
| chrX_-_55208866 | 0.24 |
ENST00000545075.1
|
MTRNR2L10
|
MT-RNR2-like 10 |
| chr17_-_71223839 | 0.23 |
ENST00000579872.1
ENST00000580032.1 |
FAM104A
|
family with sequence similarity 104, member A |
| chr5_-_64064508 | 0.22 |
ENST00000513458.4
|
SREK1IP1
|
SREK1-interacting protein 1 |
| chr8_+_128426535 | 0.22 |
ENST00000465342.2
|
POU5F1B
|
POU class 5 homeobox 1B |
| chr18_+_68002675 | 0.21 |
ENST00000584919.1
|
RP11-41O4.1
|
Uncharacterized protein |
| chr19_+_52076425 | 0.21 |
ENST00000436511.2
|
ZNF175
|
zinc finger protein 175 |
| chr1_-_150669604 | 0.21 |
ENST00000427665.1
ENST00000540514.1 |
GOLPH3L
|
golgi phosphoprotein 3-like |
| chr11_-_6633799 | 0.21 |
ENST00000299424.4
|
TAF10
|
TAF10 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 30kDa |
| chr10_+_70847852 | 0.21 |
ENST00000242465.3
|
SRGN
|
serglycin |
| chr21_-_39705286 | 0.21 |
ENST00000414189.1
|
AP001422.3
|
AP001422.3 |
| chr1_+_67632083 | 0.21 |
ENST00000347310.5
ENST00000371002.1 |
IL23R
|
interleukin 23 receptor |
| chr16_+_15528332 | 0.21 |
ENST00000566490.1
|
C16orf45
|
chromosome 16 open reading frame 45 |
| chr1_+_41448820 | 0.20 |
ENST00000372616.1
|
CTPS1
|
CTP synthase 1 |
| chr17_-_6554877 | 0.20 |
ENST00000225728.3
ENST00000575197.1 |
MED31
|
mediator complex subunit 31 |
| chr6_+_123317116 | 0.20 |
ENST00000275162.5
|
CLVS2
|
clavesin 2 |
| chr12_+_40787194 | 0.20 |
ENST00000425730.2
ENST00000454784.4 |
MUC19
|
mucin 19, oligomeric |
| chr19_+_54135310 | 0.20 |
ENST00000376650.1
|
DPRX
|
divergent-paired related homeobox |
| chr6_-_49712147 | 0.20 |
ENST00000433368.2
ENST00000354620.4 |
CRISP3
|
cysteine-rich secretory protein 3 |
| chr3_+_106959552 | 0.19 |
ENST00000473550.1
|
LINC00883
|
long intergenic non-protein coding RNA 883 |
| chr6_-_74161977 | 0.19 |
ENST00000370318.1
ENST00000370315.3 |
MB21D1
|
Mab-21 domain containing 1 |
| chr12_-_8218997 | 0.19 |
ENST00000307637.4
|
C3AR1
|
complement component 3a receptor 1 |
| chr9_-_34381511 | 0.19 |
ENST00000379124.1
|
C9orf24
|
chromosome 9 open reading frame 24 |
| chr3_+_107602030 | 0.19 |
ENST00000494231.1
|
LINC00636
|
long intergenic non-protein coding RNA 636 |
| chr12_-_96390108 | 0.19 |
ENST00000538703.1
ENST00000261208.3 |
HAL
|
histidine ammonia-lyase |
| chr6_+_24357131 | 0.19 |
ENST00000274766.1
|
KAAG1
|
kidney associated antigen 1 |
| chr3_-_106959424 | 0.19 |
ENST00000607801.1
ENST00000479612.2 ENST00000484698.1 ENST00000477210.2 ENST00000473636.1 |
LINC00882
|
long intergenic non-protein coding RNA 882 |
| chr2_-_70520832 | 0.19 |
ENST00000454893.1
ENST00000272348.2 |
SNRPG
|
small nuclear ribonucleoprotein polypeptide G |
| chr7_+_13141010 | 0.19 |
ENST00000443947.1
|
AC011288.2
|
AC011288.2 |
| chr1_-_213020991 | 0.19 |
ENST00000332912.3
|
C1orf227
|
chromosome 1 open reading frame 227 |
| chr7_-_7782204 | 0.18 |
ENST00000418534.2
|
AC007161.5
|
AC007161.5 |
| chr4_+_76871883 | 0.18 |
ENST00000599764.1
|
AC110615.1
|
Uncharacterized protein |
| chr10_+_90562705 | 0.18 |
ENST00000539337.1
|
LIPM
|
lipase, family member M |
| chr20_-_5426380 | 0.18 |
ENST00000609252.1
ENST00000422352.1 |
LINC00658
|
long intergenic non-protein coding RNA 658 |
| chr6_-_49792495 | 0.18 |
ENST00000430821.1
|
RP11-719J20.1
|
RP11-719J20.1 |
| chr10_+_114710516 | 0.18 |
ENST00000542695.1
ENST00000346198.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chrX_+_36053908 | 0.18 |
ENST00000378660.2
|
CHDC2
|
calponin homology domain containing 2 |
| chr15_-_52404921 | 0.18 |
ENST00000561198.1
ENST00000260442.3 |
BCL2L10
|
BCL2-like 10 (apoptosis facilitator) |
| chr10_+_81891416 | 0.18 |
ENST00000372270.2
|
PLAC9
|
placenta-specific 9 |
| chr5_+_140220769 | 0.17 |
ENST00000531613.1
ENST00000378123.3 |
PCDHA8
|
protocadherin alpha 8 |
| chr5_-_55412774 | 0.17 |
ENST00000434982.2
|
ANKRD55
|
ankyrin repeat domain 55 |
| chr3_+_130279178 | 0.17 |
ENST00000358511.6
ENST00000453409.2 |
COL6A6
|
collagen, type VI, alpha 6 |
| chr6_-_49712123 | 0.17 |
ENST00000263045.4
|
CRISP3
|
cysteine-rich secretory protein 3 |
| chr4_-_90756769 | 0.17 |
ENST00000345009.4
ENST00000505199.1 ENST00000502987.1 |
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr2_+_12246664 | 0.17 |
ENST00000449986.1
|
AC096559.1
|
AC096559.1 |
| chr10_+_78078088 | 0.17 |
ENST00000496424.2
|
C10orf11
|
chromosome 10 open reading frame 11 |
| chr11_+_1092184 | 0.17 |
ENST00000361558.6
|
MUC2
|
mucin 2, oligomeric mucus/gel-forming |
| chr4_+_130017268 | 0.17 |
ENST00000425929.1
ENST00000508673.1 ENST00000508622.1 |
C4orf33
|
chromosome 4 open reading frame 33 |
| chr4_+_56212270 | 0.16 |
ENST00000264228.4
|
SRD5A3
|
steroid 5 alpha-reductase 3 |
| chr17_-_6554747 | 0.16 |
ENST00000574128.1
|
MED31
|
mediator complex subunit 31 |
| chrX_+_36254051 | 0.16 |
ENST00000378657.4
|
CXorf30
|
chromosome X open reading frame 30 |
| chr12_+_8995832 | 0.16 |
ENST00000541459.1
|
A2ML1
|
alpha-2-macroglobulin-like 1 |
| chr5_-_39270725 | 0.16 |
ENST00000512138.1
ENST00000512982.1 ENST00000540520.1 |
FYB
|
FYN binding protein |
| chr10_+_47746929 | 0.16 |
ENST00000340243.6
ENST00000374277.5 ENST00000449464.2 ENST00000538825.1 ENST00000335083.5 |
ANXA8L2
AL603965.1
|
annexin A8-like 2 Protein LOC100996760 |
| chr7_+_116593292 | 0.16 |
ENST00000393446.2
ENST00000265437.5 ENST00000393451.3 |
ST7
|
suppression of tumorigenicity 7 |
| chr3_+_101546827 | 0.16 |
ENST00000461724.1
ENST00000483180.1 ENST00000394054.2 |
NFKBIZ
|
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
| chr8_-_28347737 | 0.16 |
ENST00000517673.1
ENST00000518734.1 ENST00000346498.2 ENST00000380254.2 |
FBXO16
|
F-box protein 16 |
| chr19_-_21950362 | 0.15 |
ENST00000358296.6
|
ZNF100
|
zinc finger protein 100 |
| chr7_+_116593536 | 0.15 |
ENST00000417919.1
|
ST7
|
suppression of tumorigenicity 7 |
| chr3_+_171561127 | 0.15 |
ENST00000334567.5
ENST00000450693.1 |
TMEM212
|
transmembrane protein 212 |
| chr2_+_219646462 | 0.15 |
ENST00000258415.4
|
CYP27A1
|
cytochrome P450, family 27, subfamily A, polypeptide 1 |
| chr11_-_119249805 | 0.15 |
ENST00000527843.1
|
USP2
|
ubiquitin specific peptidase 2 |
| chr10_-_126716459 | 0.15 |
ENST00000309035.6
|
CTBP2
|
C-terminal binding protein 2 |
| chr4_+_55095428 | 0.15 |
ENST00000508170.1
ENST00000512143.1 |
PDGFRA
|
platelet-derived growth factor receptor, alpha polypeptide |
| chr2_-_118943930 | 0.15 |
ENST00000449075.1
ENST00000414886.1 ENST00000449819.1 |
AC093901.1
|
AC093901.1 |
| chr10_-_21661870 | 0.15 |
ENST00000433460.1
|
RP11-275N1.1
|
RP11-275N1.1 |
| chr6_+_126221034 | 0.15 |
ENST00000433571.1
|
NCOA7
|
nuclear receptor coactivator 7 |
| chr12_-_91546926 | 0.15 |
ENST00000550758.1
|
DCN
|
decorin |
| chr7_+_107220422 | 0.15 |
ENST00000005259.4
|
BCAP29
|
B-cell receptor-associated protein 29 |
| chr3_-_150421752 | 0.14 |
ENST00000498386.1
|
FAM194A
|
family with sequence similarity 194, member A |
| chr19_-_54824344 | 0.14 |
ENST00000346508.3
ENST00000446712.3 ENST00000432233.3 ENST00000301219.3 |
LILRA5
|
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 5 |
| chr16_-_4665023 | 0.14 |
ENST00000591897.1
|
UBALD1
|
UBA-like domain containing 1 |
| chr11_-_74800799 | 0.14 |
ENST00000305159.3
|
OR2AT4
|
olfactory receptor, family 2, subfamily AT, member 4 |
| chr4_-_95264008 | 0.14 |
ENST00000295256.5
|
HPGDS
|
hematopoietic prostaglandin D synthase |
| chr3_+_52720187 | 0.14 |
ENST00000474423.1
|
GNL3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr16_+_55522536 | 0.14 |
ENST00000570283.1
|
MMP2
|
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) |
| chrX_+_36065053 | 0.14 |
ENST00000313548.4
|
CHDC2
|
calponin homology domain containing 2 |
| chr11_-_104480019 | 0.14 |
ENST00000536529.1
ENST00000545630.1 ENST00000538641.1 |
RP11-886D15.1
|
RP11-886D15.1 |
| chr5_+_68513557 | 0.13 |
ENST00000256441.4
|
MRPS36
|
mitochondrial ribosomal protein S36 |
| chr12_+_72080253 | 0.13 |
ENST00000549735.1
|
TMEM19
|
transmembrane protein 19 |
| chr1_+_144989309 | 0.13 |
ENST00000596396.1
|
AL590452.1
|
Uncharacterized protein |
| chr11_+_73661364 | 0.13 |
ENST00000339764.1
|
DNAJB13
|
DnaJ (Hsp40) homolog, subfamily B, member 13 |
| chr7_+_112262421 | 0.13 |
ENST00000453459.1
|
AC002463.3
|
AC002463.3 |
| chr9_+_112542591 | 0.13 |
ENST00000483909.1
ENST00000314527.4 ENST00000413420.1 ENST00000302798.7 ENST00000555236.1 ENST00000510514.5 |
PALM2
PALM2-AKAP2
AKAP2
|
paralemmin 2 PALM2-AKAP2 readthrough A kinase (PRKA) anchor protein 2 |
| chr12_-_57006882 | 0.13 |
ENST00000551996.1
|
BAZ2A
|
bromodomain adjacent to zinc finger domain, 2A |
| chr16_-_4664860 | 0.13 |
ENST00000587615.1
ENST00000587649.1 ENST00000590965.1 ENST00000591401.1 ENST00000283474.7 |
UBALD1
|
UBA-like domain containing 1 |
| chr6_+_46761118 | 0.13 |
ENST00000230588.4
|
MEP1A
|
meprin A, alpha (PABA peptide hydrolase) |
| chr14_+_39703112 | 0.13 |
ENST00000555143.1
ENST00000280082.3 |
MIA2
|
melanoma inhibitory activity 2 |
| chr12_+_70219052 | 0.13 |
ENST00000552032.2
ENST00000547771.2 |
MYRFL
|
myelin regulatory factor-like |
| chr18_-_50240 | 0.13 |
ENST00000573909.1
|
RP11-683L23.1
|
Tubulin beta-8 chain-like protein LOC260334 |
| chr2_+_33661382 | 0.13 |
ENST00000402538.3
|
RASGRP3
|
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chrX_-_18690210 | 0.13 |
ENST00000379984.3
|
RS1
|
retinoschisin 1 |
| chr7_-_107883678 | 0.12 |
ENST00000417701.1
|
NRCAM
|
neuronal cell adhesion molecule |
| chr6_+_151561085 | 0.12 |
ENST00000402676.2
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr15_-_41522889 | 0.12 |
ENST00000458580.2
ENST00000314992.5 ENST00000558396.1 |
EXD1
|
exonuclease 3'-5' domain containing 1 |
| chr9_-_5830768 | 0.12 |
ENST00000381506.3
|
ERMP1
|
endoplasmic reticulum metallopeptidase 1 |
| chr14_+_39583427 | 0.12 |
ENST00000308317.6
ENST00000396249.2 ENST00000250379.8 ENST00000534684.2 ENST00000527381.1 |
GEMIN2
|
gem (nuclear organelle) associated protein 2 |
| chr6_-_26199499 | 0.12 |
ENST00000377831.5
|
HIST1H3D
|
histone cluster 1, H3d |
| chr5_+_7373230 | 0.12 |
ENST00000500616.2
|
CTD-2296D1.5
|
CTD-2296D1.5 |
| chr7_-_86595190 | 0.12 |
ENST00000398276.2
ENST00000416314.1 ENST00000425689.1 |
KIAA1324L
|
KIAA1324-like |
| chr17_-_73401728 | 0.12 |
ENST00000316804.5
ENST00000316615.5 |
GRB2
|
growth factor receptor-bound protein 2 |
| chr12_-_56359802 | 0.12 |
ENST00000548803.1
ENST00000449260.2 ENST00000550447.1 ENST00000547137.1 ENST00000546543.1 ENST00000550464.1 |
PMEL
|
premelanosome protein |
| chr1_-_100643765 | 0.12 |
ENST00000370137.1
ENST00000370138.1 ENST00000342895.3 |
LRRC39
|
leucine rich repeat containing 39 |
| chr11_+_69061594 | 0.12 |
ENST00000441339.2
ENST00000308946.3 ENST00000535407.1 |
MYEOV
|
myeloma overexpressed |
| chr6_+_151561506 | 0.12 |
ENST00000253332.1
|
AKAP12
|
A kinase (PRKA) anchor protein 12 |
| chr12_-_15114603 | 0.12 |
ENST00000228945.4
|
ARHGDIB
|
Rho GDP dissociation inhibitor (GDI) beta |
| chr6_-_49712091 | 0.12 |
ENST00000371159.4
|
CRISP3
|
cysteine-rich secretory protein 3 |
| chr4_-_148605265 | 0.12 |
ENST00000541232.1
ENST00000322396.6 |
PRMT10
|
protein arginine methyltransferase 10 (putative) |
| chr12_-_48099773 | 0.11 |
ENST00000432584.3
ENST00000005386.3 |
RPAP3
|
RNA polymerase II associated protein 3 |
| chr5_-_43397184 | 0.11 |
ENST00000513525.1
|
CCL28
|
chemokine (C-C motif) ligand 28 |
| chrX_+_107288239 | 0.11 |
ENST00000217957.5
|
VSIG1
|
V-set and immunoglobulin domain containing 1 |
| chr8_+_79503458 | 0.11 |
ENST00000518467.1
|
PKIA
|
protein kinase (cAMP-dependent, catalytic) inhibitor alpha |
| chr6_+_26156551 | 0.11 |
ENST00000304218.3
|
HIST1H1E
|
histone cluster 1, H1e |
| chr15_+_65337708 | 0.11 |
ENST00000334287.2
|
SLC51B
|
solute carrier family 51, beta subunit |
| chr6_+_107077471 | 0.11 |
ENST00000369044.1
|
QRSL1
|
glutaminyl-tRNA synthase (glutamine-hydrolyzing)-like 1 |
| chr2_-_180726232 | 0.11 |
ENST00000410066.1
|
ZNF385B
|
zinc finger protein 385B |
| chr2_-_201753980 | 0.11 |
ENST00000443398.1
ENST00000286175.8 ENST00000409449.1 |
PPIL3
|
peptidylprolyl isomerase (cyclophilin)-like 3 |
| chr3_-_194072019 | 0.11 |
ENST00000429275.1
ENST00000323830.3 |
CPN2
|
carboxypeptidase N, polypeptide 2 |
| chr2_-_40739501 | 0.11 |
ENST00000403092.1
ENST00000402441.1 ENST00000448531.1 |
SLC8A1
|
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr6_-_130543958 | 0.11 |
ENST00000437477.2
ENST00000439090.2 |
SAMD3
|
sterile alpha motif domain containing 3 |
| chr4_-_47983519 | 0.11 |
ENST00000358519.4
ENST00000544810.1 ENST00000402813.3 |
CNGA1
|
cyclic nucleotide gated channel alpha 1 |
| chr11_+_4510109 | 0.11 |
ENST00000307632.3
|
OR52K1
|
olfactory receptor, family 52, subfamily K, member 1 |
| chrX_-_134186144 | 0.11 |
ENST00000370775.2
|
FAM127B
|
family with sequence similarity 127, member B |
| chr6_+_15401075 | 0.11 |
ENST00000541660.1
|
JARID2
|
jumonji, AT rich interactive domain 2 |
| chr12_+_104337515 | 0.10 |
ENST00000550595.1
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1 |
| chr12_-_96390063 | 0.10 |
ENST00000541929.1
|
HAL
|
histidine ammonia-lyase |
| chr11_-_118305921 | 0.10 |
ENST00000532619.1
|
RP11-770J1.4
|
RP11-770J1.4 |
| chr20_+_5451845 | 0.10 |
ENST00000430097.2
|
RP5-828H9.1
|
RP5-828H9.1 |
| chr12_-_10607084 | 0.10 |
ENST00000408006.3
ENST00000544822.1 ENST00000536188.1 |
KLRC1
|
killer cell lectin-like receptor subfamily C, member 1 |
| chr17_-_73401567 | 0.10 |
ENST00000392562.1
|
GRB2
|
growth factor receptor-bound protein 2 |
| chr6_-_49712072 | 0.10 |
ENST00000423399.2
|
CRISP3
|
cysteine-rich secretory protein 3 |
| chr12_+_101988627 | 0.10 |
ENST00000547405.1
ENST00000452455.2 ENST00000441232.1 ENST00000360610.2 ENST00000392934.3 ENST00000547509.1 ENST00000361685.2 ENST00000549145.1 ENST00000553190.1 |
MYBPC1
|
myosin binding protein C, slow type |
| chr10_+_97709725 | 0.10 |
ENST00000472454.2
|
RP11-248J23.6
|
Protein LOC100652732 |
| chr11_+_74459876 | 0.10 |
ENST00000299563.4
|
RNF169
|
ring finger protein 169 |
| chr7_-_120498357 | 0.10 |
ENST00000415871.1
ENST00000222747.3 ENST00000430985.1 |
TSPAN12
|
tetraspanin 12 |
| chr11_+_76745385 | 0.10 |
ENST00000533140.1
ENST00000354301.5 ENST00000528622.1 |
B3GNT6
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 6 (core 3 synthase) |
| chr13_-_41837620 | 0.10 |
ENST00000379477.1
ENST00000452359.1 ENST00000379480.4 ENST00000430347.2 |
MTRF1
|
mitochondrial translational release factor 1 |
| chr10_-_99030395 | 0.10 |
ENST00000355366.5
ENST00000371027.1 |
ARHGAP19
|
Rho GTPase activating protein 19 |
| chr8_-_62559366 | 0.10 |
ENST00000522919.1
|
ASPH
|
aspartate beta-hydroxylase |
| chr7_-_19748640 | 0.10 |
ENST00000222567.5
|
TWISTNB
|
TWIST neighbor |
| chr11_-_125648690 | 0.10 |
ENST00000436890.2
ENST00000358524.3 |
PATE2
|
prostate and testis expressed 2 |
| chr2_-_175711924 | 0.10 |
ENST00000444573.1
|
CHN1
|
chimerin 1 |
| chr3_+_11196206 | 0.09 |
ENST00000431010.2
|
HRH1
|
histamine receptor H1 |
| chr18_+_52258390 | 0.09 |
ENST00000321600.1
|
DYNAP
|
dynactin associated protein |
| chr2_+_210444748 | 0.09 |
ENST00000392194.1
|
MAP2
|
microtubule-associated protein 2 |
| chr1_-_113253977 | 0.09 |
ENST00000606505.1
ENST00000605933.1 |
RP11-426L16.10
|
Rho-related GTP-binding protein RhoC |
| chr14_+_50291993 | 0.09 |
ENST00000595378.1
|
AL627171.2
|
HCG1786899; PRO2610; Uncharacterized protein |
| chr1_+_168250194 | 0.09 |
ENST00000367821.3
|
TBX19
|
T-box 19 |
| chr3_-_87325728 | 0.09 |
ENST00000350375.2
|
POU1F1
|
POU class 1 homeobox 1 |
| chr1_+_76540386 | 0.09 |
ENST00000328299.3
|
ST6GALNAC3
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 3 |
| chr7_-_32110451 | 0.09 |
ENST00000396191.1
ENST00000396182.2 |
PDE1C
|
phosphodiesterase 1C, calmodulin-dependent 70kDa |
| chr1_+_160336851 | 0.09 |
ENST00000302101.5
|
NHLH1
|
nescient helix loop helix 1 |
| chr3_-_157221128 | 0.09 |
ENST00000392833.2
ENST00000362010.2 |
VEPH1
|
ventricular zone expressed PH domain-containing 1 |
| chrX_+_15767971 | 0.09 |
ENST00000479740.1
ENST00000454127.2 |
CA5B
|
carbonic anhydrase VB, mitochondrial |
| chr3_-_150421728 | 0.09 |
ENST00000295910.6
ENST00000491361.1 |
FAM194A
|
family with sequence similarity 194, member A |
| chr1_+_160175117 | 0.09 |
ENST00000360472.4
|
PEA15
|
phosphoprotein enriched in astrocytes 15 |
| chr14_+_62453803 | 0.08 |
ENST00000446982.2
|
SYT16
|
synaptotagmin XVI |
| chr5_+_59783540 | 0.08 |
ENST00000515734.2
|
PART1
|
prostate androgen-regulated transcript 1 (non-protein coding) |
| chr1_+_66797687 | 0.08 |
ENST00000371045.5
ENST00000531025.1 ENST00000526197.1 |
PDE4B
|
phosphodiesterase 4B, cAMP-specific |
| chr12_-_85430024 | 0.08 |
ENST00000547836.1
ENST00000532498.2 |
TSPAN19
|
tetraspanin 19 |
| chr6_+_24775153 | 0.08 |
ENST00000356509.3
ENST00000230056.3 |
GMNN
|
geminin, DNA replication inhibitor |
| chr4_+_69962185 | 0.08 |
ENST00000305231.7
|
UGT2B7
|
UDP glucuronosyltransferase 2 family, polypeptide B7 |
| chr1_-_46642154 | 0.08 |
ENST00000540385.1
|
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
| chr7_-_45957011 | 0.08 |
ENST00000417621.1
|
IGFBP3
|
insulin-like growth factor binding protein 3 |
| chr16_+_14980632 | 0.08 |
ENST00000565655.1
|
NOMO1
|
NODAL modulator 1 |
| chr19_+_30156351 | 0.08 |
ENST00000592810.1
|
PLEKHF1
|
pleckstrin homology domain containing, family F (with FYVE domain) member 1 |
| chr2_+_171034646 | 0.08 |
ENST00000409044.3
ENST00000408978.4 |
MYO3B
|
myosin IIIB |
Network of associatons between targets according to the STRING database.
First level regulatory network of ARID5A
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0072616 | interleukin-18 secretion(GO:0072616) |
| 0.1 | 0.5 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.1 | 0.6 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.1 | 0.7 | GO:0019557 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.1 | 0.3 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.1 | 0.7 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.1 | 0.3 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.1 | 0.2 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.1 | 0.9 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.6 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.2 | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.0 | 0.2 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.0 | 0.2 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.1 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.1 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.0 | 0.2 | GO:0051585 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.0 | 0.3 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.0 | 0.2 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.0 | 0.1 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.0 | 0.2 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.0 | 0.1 | GO:0060151 | peroxisome localization(GO:0060151) microtubule-based peroxisome localization(GO:0060152) |
| 0.0 | 0.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.2 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) myoblast fate commitment(GO:0048625) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.4 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.0 | 0.1 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.1 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.3 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.1 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.1 | GO:0021503 | neural fold bending(GO:0021503) |
| 0.0 | 0.2 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.1 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.0 | 0.1 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.0 | 0.1 | GO:0098507 | polynucleotide 5' dephosphorylation(GO:0098507) |
| 0.0 | 0.1 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.0 | 0.2 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.2 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.0 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.1 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.0 | 0.0 | GO:0043449 | cellular alkene metabolic process(GO:0043449) |
| 0.0 | 0.1 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.0 | 0.1 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.1 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.2 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.0 | 0.3 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.1 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) |
| 0.0 | 0.2 | GO:0070365 | hepatocyte differentiation(GO:0070365) |
| 0.0 | 0.1 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.1 | GO:0007402 | ganglion mother cell fate determination(GO:0007402) |
| 0.0 | 0.3 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.3 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.1 | GO:0072100 | apoptotic process involved in endocardial cushion morphogenesis(GO:0003277) intermediate mesoderm morphogenesis(GO:0048390) intermediate mesoderm formation(GO:0048391) intermediate mesodermal cell differentiation(GO:0048392) regulation of cardiac muscle fiber development(GO:0055018) positive regulation of cardiac muscle fiber development(GO:0055020) bud dilation involved in lung branching(GO:0060503) BMP signaling pathway involved in ureter morphogenesis(GO:0061149) renal system segmentation(GO:0061150) BMP signaling pathway involved in renal system segmentation(GO:0061151) pulmonary artery endothelial tube morphogenesis(GO:0061155) regulation of transcription from RNA polymerase II promoter involved in mesonephros development(GO:0061216) BMP signaling pathway involved in nephric duct formation(GO:0071893) negative regulation of branch elongation involved in ureteric bud branching(GO:0072096) negative regulation of branch elongation involved in ureteric bud branching by BMP signaling pathway(GO:0072097) anterior/posterior pattern specification involved in ureteric bud development(GO:0072099) specification of ureteric bud anterior/posterior symmetry(GO:0072100) specification of ureteric bud anterior/posterior symmetry by BMP signaling pathway(GO:0072101) ureter epithelial cell differentiation(GO:0072192) negative regulation of mesenchymal cell proliferation involved in ureter development(GO:0072200) regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901963) positive regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901964) cardiac jelly development(GO:1905072) regulation of metanephric S-shaped body morphogenesis(GO:2000004) negative regulation of metanephric S-shaped body morphogenesis(GO:2000005) regulation of metanephric comma-shaped body morphogenesis(GO:2000006) negative regulation of metanephric comma-shaped body morphogenesis(GO:2000007) |
| 0.0 | 0.1 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.0 | 0.1 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.1 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 0.0 | 0.1 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.4 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.1 | GO:1901078 | negative regulation of relaxation of muscle(GO:1901078) negative regulation of relaxation of cardiac muscle(GO:1901898) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0070703 | inner mucus layer(GO:0070702) outer mucus layer(GO:0070703) |
| 0.0 | 0.4 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.3 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.4 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.2 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 0.1 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.2 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.1 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 0.2 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 0.1 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.4 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.0 | 0.2 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.1 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0097635 | extrinsic component of autophagosome membrane(GO:0097635) |
| 0.0 | 0.3 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.4 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.0 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.1 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 0.1 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.5 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.2 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.1 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0004397 | histidine ammonia-lyase activity(GO:0004397) |
| 0.1 | 0.6 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.1 | 0.7 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.1 | 0.4 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.1 | 0.3 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.3 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.1 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.0 | 0.6 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.2 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.2 | GO:0042019 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 0.2 | GO:0033765 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 0.2 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.1 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.1 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.0 | 0.2 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.0 | 0.1 | GO:0047223 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,3-N-acetylglucosaminyltransferase activity(GO:0047223) |
| 0.0 | 0.1 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.0 | 0.5 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.1 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.6 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.2 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.1 | GO:0032558 | adenyl deoxyribonucleotide binding(GO:0032558) dATP binding(GO:0032564) |
| 0.0 | 0.1 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.1 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.0 | 0.1 | GO:0004651 | polynucleotide 5'-phosphatase activity(GO:0004651) |
| 0.0 | 0.2 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.1 | GO:0016429 | tRNA (adenine) methyltransferase activity(GO:0016426) tRNA (adenine-N1-)-methyltransferase activity(GO:0016429) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.2 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.4 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.2 | GO:0005168 | neurotrophin TRKA receptor binding(GO:0005168) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 1.0 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.2 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.5 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.7 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 1.2 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.2 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |