Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
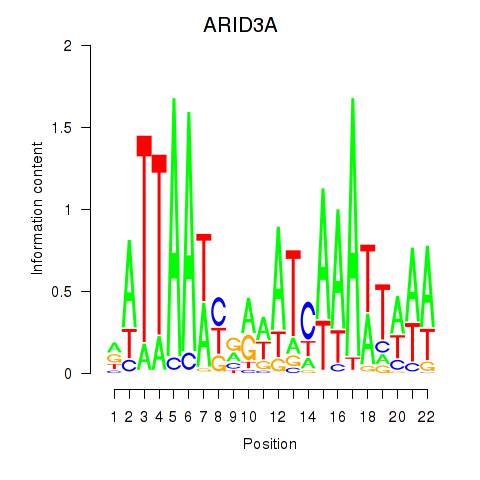
Results for ARID3A
Z-value: 0.19
Transcription factors associated with ARID3A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ARID3A
|
ENSG00000116017.6 | AT-rich interaction domain 3A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ARID3A | hg19_v2_chr19_+_926000_926046 | -0.03 | 9.7e-01 | Click! |
Activity profile of ARID3A motif
Sorted Z-values of ARID3A motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_67351019 | 0.18 |
ENST00000398606.3
|
GSTP1
|
glutathione S-transferase pi 1 |
| chr11_+_67351213 | 0.17 |
ENST00000398603.1
|
GSTP1
|
glutathione S-transferase pi 1 |
| chr4_+_113568207 | 0.15 |
ENST00000511529.1
|
LARP7
|
La ribonucleoprotein domain family, member 7 |
| chr1_+_241695670 | 0.14 |
ENST00000366557.4
|
KMO
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr3_-_139396801 | 0.13 |
ENST00000413939.2
ENST00000339837.5 ENST00000512391.1 |
NMNAT3
|
nicotinamide nucleotide adenylyltransferase 3 |
| chr6_+_26087509 | 0.13 |
ENST00000397022.3
ENST00000353147.5 ENST00000352392.4 ENST00000349999.4 ENST00000317896.7 ENST00000357618.5 ENST00000470149.1 ENST00000336625.8 ENST00000461397.1 ENST00000488199.1 |
HFE
|
hemochromatosis |
| chr9_-_69229650 | 0.11 |
ENST00000416428.1
|
CBWD6
|
COBW domain containing 6 |
| chr15_-_31521567 | 0.11 |
ENST00000560812.1
ENST00000559853.1 ENST00000558109.1 |
RP11-16E12.2
|
RP11-16E12.2 |
| chr11_-_86383650 | 0.11 |
ENST00000526944.1
ENST00000530335.1 ENST00000543262.1 ENST00000524826.1 |
ME3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial |
| chr1_+_229440129 | 0.11 |
ENST00000366688.3
|
SPHAR
|
S-phase response (cyclin related) |
| chr3_-_139396853 | 0.11 |
ENST00000406164.1
ENST00000406824.1 |
NMNAT3
|
nicotinamide nucleotide adenylyltransferase 3 |
| chr12_+_56114151 | 0.10 |
ENST00000547072.1
ENST00000552930.1 ENST00000257895.5 |
RDH5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr12_-_68696652 | 0.10 |
ENST00000539972.1
|
MDM1
|
Mdm1 nuclear protein homolog (mouse) |
| chr19_-_48613820 | 0.10 |
ENST00000596352.1
|
PLA2G4C
|
phospholipase A2, group IVC (cytosolic, calcium-independent) |
| chr8_+_74903580 | 0.10 |
ENST00000284818.2
ENST00000518893.1 |
LY96
|
lymphocyte antigen 96 |
| chr3_+_118892411 | 0.09 |
ENST00000479520.1
ENST00000494855.1 |
UPK1B
|
uroplakin 1B |
| chr3_+_118892362 | 0.09 |
ENST00000497685.1
ENST00000264234.3 |
UPK1B
|
uroplakin 1B |
| chr12_+_133757995 | 0.09 |
ENST00000536435.2
ENST00000228289.5 ENST00000541211.2 ENST00000500625.3 ENST00000539248.2 ENST00000542711.2 ENST00000536899.2 ENST00000542986.2 ENST00000537565.1 ENST00000541975.2 |
ZNF268
|
zinc finger protein 268 |
| chr4_+_141264597 | 0.09 |
ENST00000338517.4
ENST00000394203.3 ENST00000506322.1 |
SCOC
|
short coiled-coil protein |
| chr12_+_56114189 | 0.09 |
ENST00000548082.1
|
RDH5
|
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr5_-_10308125 | 0.09 |
ENST00000296658.3
|
CMBL
|
carboxymethylenebutenolidase homolog (Pseudomonas) |
| chr12_+_133758115 | 0.08 |
ENST00000541009.2
ENST00000592241.1 |
ZNF268
|
zinc finger protein 268 |
| chr10_+_62538089 | 0.08 |
ENST00000519078.2
ENST00000395284.3 ENST00000316629.4 |
CDK1
|
cyclin-dependent kinase 1 |
| chr1_+_207943667 | 0.08 |
ENST00000462968.2
|
CD46
|
CD46 molecule, complement regulatory protein |
| chr7_+_6617039 | 0.08 |
ENST00000405731.3
ENST00000396713.2 ENST00000396707.2 ENST00000335965.6 ENST00000396709.1 ENST00000483589.1 ENST00000396706.2 |
ZDHHC4
|
zinc finger, DHHC-type containing 4 |
| chr6_+_26087646 | 0.08 |
ENST00000309234.6
|
HFE
|
hemochromatosis |
| chr1_+_145438469 | 0.08 |
ENST00000369317.4
|
TXNIP
|
thioredoxin interacting protein |
| chr11_+_129936836 | 0.08 |
ENST00000597197.1
|
AP003041.2
|
Uncharacterized protein |
| chr16_-_21289627 | 0.08 |
ENST00000396023.2
ENST00000415987.2 |
CRYM
|
crystallin, mu |
| chr19_+_49199209 | 0.07 |
ENST00000522966.1
ENST00000425340.2 ENST00000391876.4 |
FUT2
|
fucosyltransferase 2 (secretor status included) |
| chr15_-_45670924 | 0.07 |
ENST00000396659.3
|
GATM
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr12_-_74686314 | 0.07 |
ENST00000551210.1
ENST00000515416.2 ENST00000549905.1 |
RP11-81H3.2
|
RP11-81H3.2 |
| chr17_+_38516907 | 0.07 |
ENST00000578774.1
|
CTD-2267D19.3
|
Uncharacterized protein |
| chr19_-_13900972 | 0.07 |
ENST00000397557.1
|
AC008686.1
|
Uncharacterized protein |
| chr4_-_110723335 | 0.07 |
ENST00000394634.2
|
CFI
|
complement factor I |
| chrX_-_63005405 | 0.07 |
ENST00000374878.1
ENST00000437457.2 |
ARHGEF9
|
Cdc42 guanine nucleotide exchange factor (GEF) 9 |
| chr13_+_98612446 | 0.07 |
ENST00000496368.1
ENST00000421861.2 ENST00000357602.3 |
IPO5
|
importin 5 |
| chr11_-_86383370 | 0.07 |
ENST00000526834.1
ENST00000359636.2 |
ME3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial |
| chr11_+_327171 | 0.06 |
ENST00000534483.1
ENST00000524824.1 ENST00000531076.1 |
RP11-326C3.12
|
RP11-326C3.12 |
| chr1_-_235098935 | 0.06 |
ENST00000423175.1
|
RP11-443B7.1
|
RP11-443B7.1 |
| chrX_+_80457442 | 0.06 |
ENST00000373212.5
|
SH3BGRL
|
SH3 domain binding glutamic acid-rich protein like |
| chr11_-_104035088 | 0.06 |
ENST00000302251.5
|
PDGFD
|
platelet derived growth factor D |
| chr2_-_176867534 | 0.06 |
ENST00000445472.1
|
KIAA1715
|
KIAA1715 |
| chr7_+_65552756 | 0.06 |
ENST00000450043.1
|
AC068533.7
|
AC068533.7 |
| chr3_-_121379739 | 0.06 |
ENST00000428394.2
ENST00000314583.3 |
HCLS1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr3_+_186158169 | 0.06 |
ENST00000435548.1
ENST00000421006.1 |
RP11-78H24.1
|
RP11-78H24.1 |
| chr5_+_96038476 | 0.06 |
ENST00000511049.1
ENST00000309190.5 ENST00000510156.1 ENST00000509903.1 ENST00000511782.1 ENST00000504465.1 |
CAST
|
calpastatin |
| chr4_-_110723194 | 0.06 |
ENST00000394635.3
|
CFI
|
complement factor I |
| chr15_-_37390482 | 0.06 |
ENST00000559085.1
ENST00000397624.3 |
MEIS2
|
Meis homeobox 2 |
| chr13_+_110958124 | 0.06 |
ENST00000400163.2
|
COL4A2
|
collagen, type IV, alpha 2 |
| chr10_-_69597810 | 0.06 |
ENST00000483798.2
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr10_+_104614008 | 0.05 |
ENST00000369883.3
|
C10orf32
|
chromosome 10 open reading frame 32 |
| chr2_-_191115229 | 0.05 |
ENST00000409820.2
ENST00000410045.1 |
HIBCH
|
3-hydroxyisobutyryl-CoA hydrolase |
| chr14_+_39734482 | 0.05 |
ENST00000554392.1
ENST00000555716.1 ENST00000341749.3 ENST00000557038.1 |
CTAGE5
|
CTAGE family, member 5 |
| chr16_-_46649905 | 0.05 |
ENST00000569702.1
|
SHCBP1
|
SHC SH2-domain binding protein 1 |
| chr10_-_69597915 | 0.05 |
ENST00000225171.2
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr6_-_136847099 | 0.05 |
ENST00000438100.2
|
MAP7
|
microtubule-associated protein 7 |
| chr6_-_100016678 | 0.05 |
ENST00000523799.1
ENST00000520429.1 |
CCNC
|
cyclin C |
| chr5_+_68860949 | 0.05 |
ENST00000507595.1
|
GTF2H2C
|
general transcription factor IIH, polypeptide 2C |
| chr8_-_6115044 | 0.05 |
ENST00000519555.1
|
RP11-124B13.1
|
RP11-124B13.1 |
| chr12_+_124457746 | 0.05 |
ENST00000392404.3
ENST00000538932.2 ENST00000337815.4 ENST00000540762.2 |
ZNF664
FAM101A
|
zinc finger protein 664 family with sequence similarity 101, member A |
| chr14_+_97263641 | 0.05 |
ENST00000216639.3
|
VRK1
|
vaccinia related kinase 1 |
| chr17_+_41005283 | 0.05 |
ENST00000592999.1
|
AOC3
|
amine oxidase, copper containing 3 |
| chr1_-_247267580 | 0.05 |
ENST00000366501.1
ENST00000366500.1 ENST00000476158.2 ENST00000448299.2 ENST00000358785.4 ENST00000343381.6 |
ZNF669
|
zinc finger protein 669 |
| chr19_-_55836697 | 0.05 |
ENST00000438693.1
ENST00000591570.1 |
TMEM150B
|
transmembrane protein 150B |
| chr2_+_183582774 | 0.05 |
ENST00000537515.1
|
DNAJC10
|
DnaJ (Hsp40) homolog, subfamily C, member 10 |
| chr7_-_124569991 | 0.05 |
ENST00000446993.1
ENST00000357628.3 ENST00000393329.1 |
POT1
|
protection of telomeres 1 |
| chr6_-_108278456 | 0.04 |
ENST00000429168.1
|
SEC63
|
SEC63 homolog (S. cerevisiae) |
| chr6_-_100016527 | 0.04 |
ENST00000523985.1
ENST00000518714.1 ENST00000520371.1 |
CCNC
|
cyclin C |
| chr10_+_104613980 | 0.04 |
ENST00000339834.5
|
C10orf32
|
chromosome 10 open reading frame 32 |
| chr3_+_142442841 | 0.04 |
ENST00000476941.1
ENST00000273482.6 |
TRPC1
|
transient receptor potential cation channel, subfamily C, member 1 |
| chr19_-_55836669 | 0.04 |
ENST00000326652.4
|
TMEM150B
|
transmembrane protein 150B |
| chr4_-_100242549 | 0.04 |
ENST00000305046.8
ENST00000394887.3 |
ADH1B
|
alcohol dehydrogenase 1B (class I), beta polypeptide |
| chr13_-_20110902 | 0.04 |
ENST00000390680.2
ENST00000382977.4 ENST00000382975.4 ENST00000457266.2 |
TPTE2
|
transmembrane phosphoinositide 3-phosphatase and tensin homolog 2 |
| chr1_+_156095951 | 0.04 |
ENST00000448611.2
ENST00000368297.1 |
LMNA
|
lamin A/C |
| chr6_-_30181156 | 0.04 |
ENST00000418026.1
ENST00000416596.1 ENST00000453195.1 |
TRIM26
|
tripartite motif containing 26 |
| chr12_-_39734783 | 0.04 |
ENST00000552961.1
|
KIF21A
|
kinesin family member 21A |
| chr14_+_61449076 | 0.04 |
ENST00000526105.1
|
SLC38A6
|
solute carrier family 38, member 6 |
| chr12_+_72061563 | 0.04 |
ENST00000551238.1
|
THAP2
|
THAP domain containing, apoptosis associated protein 2 |
| chr10_-_46620012 | 0.04 |
ENST00000508602.1
ENST00000374339.3 ENST00000502254.1 ENST00000437863.1 ENST00000374342.2 ENST00000395722.3 |
PTPN20A
|
protein tyrosine phosphatase, non-receptor type 20A |
| chr17_-_56494908 | 0.04 |
ENST00000577716.1
|
RNF43
|
ring finger protein 43 |
| chr3_-_139396560 | 0.04 |
ENST00000514703.1
ENST00000511444.1 |
NMNAT3
|
nicotinamide nucleotide adenylyltransferase 3 |
| chr6_-_30181133 | 0.04 |
ENST00000454678.2
ENST00000434785.1 |
TRIM26
|
tripartite motif containing 26 |
| chr12_-_102591604 | 0.04 |
ENST00000329406.4
|
PMCH
|
pro-melanin-concentrating hormone |
| chr12_+_53399942 | 0.04 |
ENST00000262056.9
|
EIF4B
|
eukaryotic translation initiation factor 4B |
| chr14_-_75518129 | 0.04 |
ENST00000556257.1
ENST00000557648.1 ENST00000553263.1 ENST00000355774.2 ENST00000380968.2 ENST00000238662.7 |
MLH3
|
mutL homolog 3 |
| chr2_-_160473114 | 0.04 |
ENST00000392783.2
|
BAZ2B
|
bromodomain adjacent to zinc finger domain, 2B |
| chr9_-_27005686 | 0.04 |
ENST00000380055.5
|
LRRC19
|
leucine rich repeat containing 19 |
| chr17_-_56494882 | 0.04 |
ENST00000584437.1
|
RNF43
|
ring finger protein 43 |
| chr9_+_42671887 | 0.04 |
ENST00000456520.1
ENST00000377391.3 |
CBWD7
|
COBW domain containing 7 |
| chr6_-_52860171 | 0.04 |
ENST00000370963.4
|
GSTA4
|
glutathione S-transferase alpha 4 |
| chr15_-_54051831 | 0.04 |
ENST00000557913.1
ENST00000360509.5 |
WDR72
|
WD repeat domain 72 |
| chr12_-_53601000 | 0.04 |
ENST00000338737.4
ENST00000549086.2 |
ITGB7
|
integrin, beta 7 |
| chr22_+_18721427 | 0.04 |
ENST00000342888.3
|
AC008132.1
|
Uncharacterized protein |
| chr3_+_39424828 | 0.04 |
ENST00000273158.4
ENST00000431510.1 |
SLC25A38
|
solute carrier family 25, member 38 |
| chr1_-_207206092 | 0.04 |
ENST00000359470.5
ENST00000461135.2 |
C1orf116
|
chromosome 1 open reading frame 116 |
| chr2_-_3521518 | 0.04 |
ENST00000382093.5
|
ADI1
|
acireductone dioxygenase 1 |
| chr5_+_68513622 | 0.04 |
ENST00000512880.1
ENST00000602380.1 |
MRPS36
|
mitochondrial ribosomal protein S36 |
| chr5_-_42887494 | 0.04 |
ENST00000514218.1
|
SEPP1
|
selenoprotein P, plasma, 1 |
| chr13_-_79233314 | 0.04 |
ENST00000282003.6
|
RNF219
|
ring finger protein 219 |
| chr1_+_234509413 | 0.04 |
ENST00000366613.1
ENST00000366612.1 |
COA6
|
cytochrome c oxidase assembly factor 6 homolog (S. cerevisiae) |
| chr2_-_85555385 | 0.04 |
ENST00000377386.3
|
TGOLN2
|
trans-golgi network protein 2 |
| chr14_+_24099318 | 0.04 |
ENST00000432832.2
|
DHRS2
|
dehydrogenase/reductase (SDR family) member 2 |
| chr12_-_120765565 | 0.03 |
ENST00000423423.3
ENST00000308366.4 |
PLA2G1B
|
phospholipase A2, group IB (pancreas) |
| chr4_+_109571740 | 0.03 |
ENST00000361564.4
|
OSTC
|
oligosaccharyltransferase complex subunit (non-catalytic) |
| chr1_-_55230165 | 0.03 |
ENST00000371279.3
|
PARS2
|
prolyl-tRNA synthetase 2, mitochondrial (putative) |
| chr12_+_32832134 | 0.03 |
ENST00000452533.2
|
DNM1L
|
dynamin 1-like |
| chr17_+_18684563 | 0.03 |
ENST00000476139.1
|
TVP23B
|
trans-golgi network vesicle protein 23 homolog B (S. cerevisiae) |
| chr8_+_93895865 | 0.03 |
ENST00000391681.1
|
AC117834.1
|
AC117834.1 |
| chr12_+_10658489 | 0.03 |
ENST00000538173.1
|
EIF2S3L
|
Putative eukaryotic translation initiation factor 2 subunit 3-like protein |
| chr1_+_95616933 | 0.03 |
ENST00000604203.1
|
RP11-57H12.6
|
TMEM56-RWDD3 readthrough |
| chr17_-_74351010 | 0.03 |
ENST00000435555.2
|
PRPSAP1
|
phosphoribosyl pyrophosphate synthetase-associated protein 1 |
| chr6_-_131211534 | 0.03 |
ENST00000456097.2
|
EPB41L2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr1_-_19615744 | 0.03 |
ENST00000361640.4
|
AKR7A3
|
aldo-keto reductase family 7, member A3 (aflatoxin aldehyde reductase) |
| chr10_-_69597828 | 0.03 |
ENST00000339758.7
|
DNAJC12
|
DnaJ (Hsp40) homolog, subfamily C, member 12 |
| chr2_+_190541153 | 0.03 |
ENST00000313581.4
ENST00000438402.2 ENST00000431575.2 ENST00000281412.6 |
ANKAR
|
ankyrin and armadillo repeat containing |
| chr4_-_170948361 | 0.03 |
ENST00000393702.3
|
MFAP3L
|
microfibrillar-associated protein 3-like |
| chr4_-_185275104 | 0.03 |
ENST00000317596.3
|
RP11-290F5.2
|
RP11-290F5.2 |
| chr9_-_110540419 | 0.03 |
ENST00000398726.3
|
AL162389.1
|
Uncharacterized protein |
| chr1_+_196743943 | 0.03 |
ENST00000471440.2
ENST00000391985.3 |
CFHR3
|
complement factor H-related 3 |
| chr3_-_52804872 | 0.03 |
ENST00000535191.1
ENST00000461689.1 ENST00000383721.4 ENST00000233027.5 |
NEK4
|
NIMA-related kinase 4 |
| chr18_+_29671812 | 0.03 |
ENST00000261593.3
ENST00000578914.1 |
RNF138
|
ring finger protein 138, E3 ubiquitin protein ligase |
| chrX_-_20159934 | 0.03 |
ENST00000379593.1
ENST00000379607.5 |
EIF1AX
|
eukaryotic translation initiation factor 1A, X-linked |
| chr15_+_75491203 | 0.03 |
ENST00000562637.1
|
C15orf39
|
chromosome 15 open reading frame 39 |
| chr1_+_53308398 | 0.03 |
ENST00000371528.1
|
ZYG11A
|
zyg-11 family member A, cell cycle regulator |
| chr6_+_3259122 | 0.03 |
ENST00000438998.2
ENST00000380305.4 |
PSMG4
|
proteasome (prosome, macropain) assembly chaperone 4 |
| chr15_-_75748143 | 0.03 |
ENST00000568431.1
ENST00000568309.1 ENST00000568190.1 ENST00000570115.1 ENST00000564778.1 |
SIN3A
|
SIN3 transcription regulator family member A |
| chr12_-_122879969 | 0.03 |
ENST00000540304.1
|
CLIP1
|
CAP-GLY domain containing linker protein 1 |
| chr12_+_69633317 | 0.03 |
ENST00000435070.2
|
CPSF6
|
cleavage and polyadenylation specific factor 6, 68kDa |
| chr10_-_5227096 | 0.03 |
ENST00000488756.1
ENST00000334314.3 |
AKR1CL1
|
aldo-keto reductase family 1, member C-like 1 |
| chr17_-_7531121 | 0.03 |
ENST00000573566.1
ENST00000269298.5 |
SAT2
|
spermidine/spermine N1-acetyltransferase family member 2 |
| chr1_+_211499957 | 0.03 |
ENST00000336184.2
|
TRAF5
|
TNF receptor-associated factor 5 |
| chr11_+_18154059 | 0.03 |
ENST00000531264.1
|
MRGPRX3
|
MAS-related GPR, member X3 |
| chr19_+_13001840 | 0.03 |
ENST00000222214.5
ENST00000589039.1 ENST00000591470.1 ENST00000457854.1 ENST00000422947.2 ENST00000588905.1 ENST00000587072.1 |
GCDH
|
glutaryl-CoA dehydrogenase |
| chrX_+_101854096 | 0.03 |
ENST00000246174.2
ENST00000537008.1 ENST00000541409.1 |
ARMCX5
|
armadillo repeat containing, X-linked 5 |
| chr16_+_202686 | 0.03 |
ENST00000252951.2
|
HBZ
|
hemoglobin, zeta |
| chr17_-_56494713 | 0.03 |
ENST00000407977.2
|
RNF43
|
ring finger protein 43 |
| chr3_+_12329397 | 0.03 |
ENST00000397015.2
|
PPARG
|
peroxisome proliferator-activated receptor gamma |
| chr19_-_20748614 | 0.03 |
ENST00000596797.1
|
ZNF737
|
zinc finger protein 737 |
| chr16_-_46655538 | 0.03 |
ENST00000303383.3
|
SHCBP1
|
SHC SH2-domain binding protein 1 |
| chr6_-_52859968 | 0.03 |
ENST00000370959.1
|
GSTA4
|
glutathione S-transferase alpha 4 |
| chr12_+_10658201 | 0.03 |
ENST00000322446.3
|
EIF2S3L
|
Putative eukaryotic translation initiation factor 2 subunit 3-like protein |
| chr1_-_39347255 | 0.03 |
ENST00000454994.2
ENST00000357771.3 |
GJA9
|
gap junction protein, alpha 9, 59kDa |
| chr11_+_112046190 | 0.02 |
ENST00000357685.5
ENST00000393032.2 ENST00000361053.4 |
BCO2
|
beta-carotene oxygenase 2 |
| chr12_-_11287243 | 0.02 |
ENST00000539585.1
|
TAS2R30
|
taste receptor, type 2, member 30 |
| chr18_-_51884204 | 0.02 |
ENST00000577499.1
ENST00000584040.1 ENST00000581310.1 |
STARD6
|
StAR-related lipid transfer (START) domain containing 6 |
| chrX_+_21857717 | 0.02 |
ENST00000379484.5
|
MBTPS2
|
membrane-bound transcription factor peptidase, site 2 |
| chr22_+_24198890 | 0.02 |
ENST00000345044.6
|
SLC2A11
|
solute carrier family 2 (facilitated glucose transporter), member 11 |
| chr3_+_36421826 | 0.02 |
ENST00000273183.3
|
STAC
|
SH3 and cysteine rich domain |
| chr12_+_97306295 | 0.02 |
ENST00000457368.2
|
NEDD1
|
neural precursor cell expressed, developmentally down-regulated 1 |
| chr6_-_100016492 | 0.02 |
ENST00000369217.4
ENST00000369220.4 ENST00000482541.2 |
CCNC
|
cyclin C |
| chr1_-_53387352 | 0.02 |
ENST00000541281.1
|
ECHDC2
|
enoyl CoA hydratase domain containing 2 |
| chr7_+_115862858 | 0.02 |
ENST00000393481.2
|
TES
|
testis derived transcript (3 LIM domains) |
| chr9_+_118916082 | 0.02 |
ENST00000328252.3
|
PAPPA
|
pregnancy-associated plasma protein A, pappalysin 1 |
| chrX_-_64254587 | 0.02 |
ENST00000337990.2
|
ZC4H2
|
zinc finger, C4H2 domain containing |
| chr22_-_38239808 | 0.02 |
ENST00000406423.1
ENST00000424350.1 ENST00000458278.2 |
ANKRD54
|
ankyrin repeat domain 54 |
| chr14_+_70918874 | 0.02 |
ENST00000603540.1
|
ADAM21
|
ADAM metallopeptidase domain 21 |
| chr4_-_69536346 | 0.02 |
ENST00000338206.5
|
UGT2B15
|
UDP glucuronosyltransferase 2 family, polypeptide B15 |
| chr7_-_77325545 | 0.02 |
ENST00000447009.1
ENST00000416650.1 ENST00000440088.1 ENST00000430801.1 ENST00000398043.2 |
RSBN1L-AS1
|
RSBN1L antisense RNA 1 |
| chr5_-_107703556 | 0.02 |
ENST00000496714.1
|
FBXL17
|
F-box and leucine-rich repeat protein 17 |
| chr7_-_66460563 | 0.02 |
ENST00000246868.2
|
SBDS
|
Shwachman-Bodian-Diamond syndrome |
| chr14_-_50101931 | 0.02 |
ENST00000298292.8
ENST00000406043.3 |
DNAAF2
|
dynein, axonemal, assembly factor 2 |
| chr17_+_71228740 | 0.02 |
ENST00000268942.8
ENST00000359042.2 |
C17orf80
|
chromosome 17 open reading frame 80 |
| chr18_+_51884251 | 0.02 |
ENST00000578138.1
|
C18orf54
|
chromosome 18 open reading frame 54 |
| chr4_+_159131596 | 0.02 |
ENST00000512481.1
|
TMEM144
|
transmembrane protein 144 |
| chr9_-_3489428 | 0.02 |
ENST00000451859.1
|
RFX3
|
regulatory factor X, 3 (influences HLA class II expression) |
| chr8_+_26150628 | 0.02 |
ENST00000523925.1
ENST00000315985.7 |
PPP2R2A
|
protein phosphatase 2, regulatory subunit B, alpha |
| chr4_+_71017791 | 0.02 |
ENST00000502294.1
|
C4orf40
|
chromosome 4 open reading frame 40 |
| chr17_+_41006095 | 0.02 |
ENST00000591562.1
ENST00000588033.1 |
AOC3
|
amine oxidase, copper containing 3 |
| chrY_+_16634483 | 0.02 |
ENST00000382872.1
|
NLGN4Y
|
neuroligin 4, Y-linked |
| chr6_+_111303218 | 0.02 |
ENST00000441448.2
|
RPF2
|
ribosome production factor 2 homolog (S. cerevisiae) |
| chr17_-_15466742 | 0.02 |
ENST00000584811.1
ENST00000419890.2 |
TVP23C
|
trans-golgi network vesicle protein 23 homolog C (S. cerevisiae) |
| chr4_+_100737954 | 0.02 |
ENST00000296414.7
ENST00000512369.1 |
DAPP1
|
dual adaptor of phosphotyrosine and 3-phosphoinositides |
| chr13_-_88323218 | 0.02 |
ENST00000436290.2
ENST00000453832.2 ENST00000606590.1 |
MIR4500HG
|
MIR4500 host gene (non-protein coding) |
| chr16_-_85722530 | 0.02 |
ENST00000253462.3
|
GINS2
|
GINS complex subunit 2 (Psf2 homolog) |
| chr19_-_53758094 | 0.02 |
ENST00000601828.1
ENST00000598513.1 ENST00000599012.1 ENST00000333952.4 ENST00000598806.1 |
ZNF677
|
zinc finger protein 677 |
| chr2_+_106468204 | 0.02 |
ENST00000425756.1
ENST00000393349.2 |
NCK2
|
NCK adaptor protein 2 |
| chr8_+_32579321 | 0.02 |
ENST00000522402.1
|
NRG1
|
neuregulin 1 |
| chr9_-_125590818 | 0.02 |
ENST00000259467.4
|
PDCL
|
phosducin-like |
| chr3_-_36781352 | 0.02 |
ENST00000416516.2
|
DCLK3
|
doublecortin-like kinase 3 |
| chr8_-_2585929 | 0.02 |
ENST00000519393.1
ENST00000520842.1 ENST00000520570.1 ENST00000517357.1 ENST00000517984.1 ENST00000523971.1 |
RP11-134O21.1
|
RP11-134O21.1 |
| chr10_+_7745232 | 0.02 |
ENST00000358415.4
|
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr3_-_130745403 | 0.02 |
ENST00000504725.1
ENST00000509060.1 |
ASTE1
|
asteroid homolog 1 (Drosophila) |
| chr15_-_74658519 | 0.02 |
ENST00000450547.1
ENST00000358632.4 |
CYP11A1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr2_+_183982238 | 0.02 |
ENST00000442895.2
ENST00000446612.1 ENST00000409798.1 |
NUP35
|
nucleoporin 35kDa |
| chrX_-_2882296 | 0.02 |
ENST00000438544.1
ENST00000381134.3 ENST00000545496.1 |
ARSE
|
arylsulfatase E (chondrodysplasia punctata 1) |
| chr7_+_64838712 | 0.02 |
ENST00000328747.7
ENST00000431504.1 ENST00000357512.2 |
ZNF92
|
zinc finger protein 92 |
| chr16_+_19422035 | 0.02 |
ENST00000381414.4
ENST00000396229.2 |
TMC5
|
transmembrane channel-like 5 |
| chr1_+_196743912 | 0.02 |
ENST00000367425.4
|
CFHR3
|
complement factor H-related 3 |
| chr18_+_52258390 | 0.02 |
ENST00000321600.1
|
DYNAP
|
dynactin associated protein |
| chrX_+_49091920 | 0.02 |
ENST00000376227.3
|
CCDC22
|
coiled-coil domain containing 22 |
| chr15_-_74658493 | 0.02 |
ENST00000419019.2
ENST00000569662.1 |
CYP11A1
|
cytochrome P450, family 11, subfamily A, polypeptide 1 |
| chr16_-_30064244 | 0.02 |
ENST00000571269.1
ENST00000561666.1 |
FAM57B
|
family with sequence similarity 57, member B |
| chr3_+_12329358 | 0.02 |
ENST00000309576.6
|
PPARG
|
peroxisome proliferator-activated receptor gamma |
| chr16_+_67381263 | 0.02 |
ENST00000541146.1
ENST00000563189.1 ENST00000290940.7 |
LRRC36
|
leucine rich repeat containing 36 |
| chr4_+_139694701 | 0.02 |
ENST00000502606.1
|
RP11-98O2.1
|
RP11-98O2.1 |
| chrX_+_119384607 | 0.02 |
ENST00000326624.2
ENST00000557385.1 |
ZBTB33
|
zinc finger and BTB domain containing 33 |
| chr18_+_11857439 | 0.02 |
ENST00000602628.1
|
GNAL
|
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr10_+_7745303 | 0.02 |
ENST00000429820.1
ENST00000379587.4 |
ITIH2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr22_+_20692438 | 0.02 |
ENST00000434783.3
|
FAM230A
|
family with sequence similarity 230, member A |
Network of associatons between targets according to the STRING database.
First level regulatory network of ARID3A
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.1 | 0.2 | GO:0002590 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 0.1 | 0.2 | GO:0046022 | positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.0 | 0.3 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.1 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 0.0 | 0.1 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:2000437 | monocyte extravasation(GO:0035696) regulation of monocyte extravasation(GO:2000437) |
| 0.0 | 0.2 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.0 | 0.1 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.1 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 0.0 | 0.1 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 0.0 | 0.0 | GO:0036466 | synaptic vesicle recycling via endosome(GO:0036466) |
| 0.0 | 0.0 | GO:0060382 | regulation of DNA strand elongation(GO:0060382) |
| 0.0 | 0.0 | GO:0031204 | posttranslational protein targeting to membrane, translocation(GO:0031204) |
| 0.0 | 0.0 | GO:0006433 | prolyl-tRNA aminoacylation(GO:0006433) |
| 0.0 | 0.0 | GO:0060694 | regulation of cholesterol transporter activity(GO:0060694) negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0097057 | TRAF2-GSTP1 complex(GO:0097057) |
| 0.0 | 0.2 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.1 | GO:0097125 | cyclin B1-CDK1 complex(GO:0097125) |
| 0.0 | 0.1 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.0 | 0.1 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0070026 | nitric oxide binding(GO:0070026) |
| 0.0 | 0.3 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 0.0 | 0.2 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.2 | GO:0004473 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.0 | 0.1 | GO:0015067 | amidinotransferase activity(GO:0015067) glycine amidinotransferase activity(GO:0015068) |
| 0.0 | 0.0 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.1 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 0.1 | GO:0052595 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.0 | 0.0 | GO:0008386 | cholesterol monooxygenase (side-chain-cleaving) activity(GO:0008386) |
| 0.0 | 0.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.0 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.0 | 0.0 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.0 | 0.0 | GO:0004827 | proline-tRNA ligase activity(GO:0004827) |
| 0.0 | 0.1 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.0 | 0.0 | GO:0051800 | phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |