Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
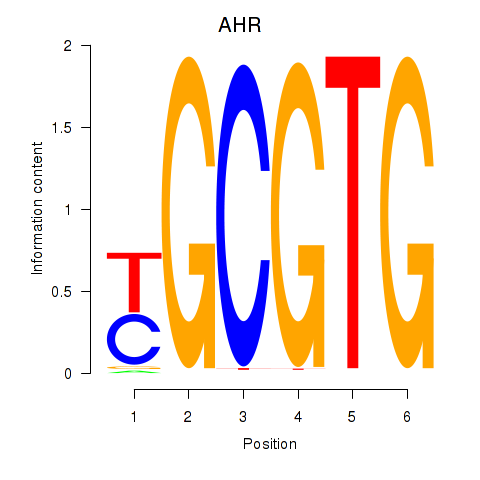
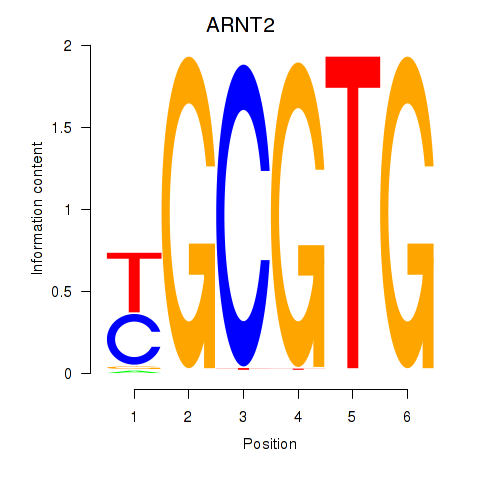
Results for AHR_ARNT2
Z-value: 0.87
Transcription factors associated with AHR_ARNT2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
AHR
|
ENSG00000106546.8 | aryl hydrocarbon receptor |
|
ARNT2
|
ENSG00000172379.14 | aryl hydrocarbon receptor nuclear translocator 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| AHR | hg19_v2_chr7_+_17338239_17338262 | 0.72 | 2.8e-01 | Click! |
| ARNT2 | hg19_v2_chr15_+_80733570_80733659 | -0.15 | 8.5e-01 | Click! |
Activity profile of AHR_ARNT2 motif
Sorted Z-values of AHR_ARNT2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_37391507 | 0.47 |
ENST00000557796.2
ENST00000397620.2 |
MEIS2
|
Meis homeobox 2 |
| chr19_-_18314799 | 0.46 |
ENST00000481914.2
|
RAB3A
|
RAB3A, member RAS oncogene family |
| chr11_-_75062730 | 0.44 |
ENST00000420843.2
ENST00000360025.3 |
ARRB1
|
arrestin, beta 1 |
| chr10_-_118885954 | 0.43 |
ENST00000392901.4
|
KIAA1598
|
KIAA1598 |
| chr11_-_75062829 | 0.42 |
ENST00000393505.4
|
ARRB1
|
arrestin, beta 1 |
| chr5_+_667759 | 0.42 |
ENST00000594226.1
|
AC026740.1
|
Uncharacterized protein |
| chr15_-_37391614 | 0.34 |
ENST00000219869.9
|
MEIS2
|
Meis homeobox 2 |
| chr14_-_23770683 | 0.34 |
ENST00000561437.1
ENST00000559942.1 ENST00000560913.1 ENST00000559314.1 ENST00000558058.1 |
PPP1R3E
|
protein phosphatase 1, regulatory subunit 3E |
| chr4_-_76598296 | 0.33 |
ENST00000395719.3
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr17_-_74533963 | 0.32 |
ENST00000293230.5
|
CYGB
|
cytoglobin |
| chr19_-_18314836 | 0.31 |
ENST00000464076.3
ENST00000222256.4 |
RAB3A
|
RAB3A, member RAS oncogene family |
| chr22_+_42372931 | 0.31 |
ENST00000328414.8
ENST00000396425.3 |
SEPT3
|
septin 3 |
| chr6_-_121655850 | 0.28 |
ENST00000422369.1
|
TBC1D32
|
TBC1 domain family, member 32 |
| chr14_-_50065882 | 0.27 |
ENST00000539688.1
|
AL139099.1
|
Full-length cDNA clone CS0DK012YO09 of HeLa cells of Homo sapiens (human); Uncharacterized protein |
| chr7_+_12250943 | 0.27 |
ENST00000442107.1
|
TMEM106B
|
transmembrane protein 106B |
| chr11_-_2292182 | 0.26 |
ENST00000331289.4
|
ASCL2
|
achaete-scute family bHLH transcription factor 2 |
| chr19_+_45445524 | 0.25 |
ENST00000591600.1
|
APOC4
|
apolipoprotein C-IV |
| chr15_+_41952591 | 0.24 |
ENST00000566718.1
ENST00000219905.7 ENST00000389936.4 ENST00000545763.1 |
MGA
|
MGA, MAX dimerization protein |
| chr5_-_81046922 | 0.24 |
ENST00000514493.1
ENST00000320672.4 |
SSBP2
|
single-stranded DNA binding protein 2 |
| chr9_+_111696664 | 0.22 |
ENST00000374624.3
ENST00000445175.1 |
FAM206A
|
family with sequence similarity 206, member A |
| chr1_+_145438469 | 0.22 |
ENST00000369317.4
|
TXNIP
|
thioredoxin interacting protein |
| chr5_-_81046904 | 0.22 |
ENST00000515395.1
|
SSBP2
|
single-stranded DNA binding protein 2 |
| chr11_-_86383650 | 0.22 |
ENST00000526944.1
ENST00000530335.1 ENST00000543262.1 ENST00000524826.1 |
ME3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial |
| chr12_+_77158021 | 0.21 |
ENST00000550876.1
|
ZDHHC17
|
zinc finger, DHHC-type containing 17 |
| chr3_+_49449636 | 0.21 |
ENST00000273590.3
|
TCTA
|
T-cell leukemia translocation altered |
| chr21_+_17961006 | 0.21 |
ENST00000602323.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr20_+_48909240 | 0.20 |
ENST00000371639.3
|
RP11-290F20.1
|
RP11-290F20.1 |
| chr5_+_60628074 | 0.20 |
ENST00000252744.5
|
ZSWIM6
|
zinc finger, SWIM-type containing 6 |
| chr17_-_77179487 | 0.20 |
ENST00000580508.1
|
RBFOX3
|
RNA binding protein, fox-1 homolog (C. elegans) 3 |
| chr6_-_31869769 | 0.20 |
ENST00000375527.2
|
ZBTB12
|
zinc finger and BTB domain containing 12 |
| chr5_-_142783175 | 0.20 |
ENST00000231509.3
ENST00000394464.2 |
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr12_+_133707161 | 0.20 |
ENST00000538918.1
ENST00000540609.1 ENST00000248211.6 ENST00000543271.1 ENST00000536877.1 |
ZNF10
|
zinc finger protein 10 |
| chr22_+_42372764 | 0.19 |
ENST00000396426.3
ENST00000406029.1 |
SEPT3
|
septin 3 |
| chr10_-_90751038 | 0.19 |
ENST00000458159.1
ENST00000415557.1 ENST00000458208.1 |
ACTA2
|
actin, alpha 2, smooth muscle, aorta |
| chr17_-_27230035 | 0.19 |
ENST00000378895.4
ENST00000394901.3 |
DHRS13
|
dehydrogenase/reductase (SDR family) member 13 |
| chr6_-_121655593 | 0.19 |
ENST00000398212.2
|
TBC1D32
|
TBC1 domain family, member 32 |
| chr6_-_10412600 | 0.19 |
ENST00000379608.3
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
| chr6_-_79787902 | 0.19 |
ENST00000275034.4
|
PHIP
|
pleckstrin homology domain interacting protein |
| chr20_+_34680620 | 0.18 |
ENST00000430276.1
ENST00000373950.2 ENST00000452261.1 |
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr6_+_26087509 | 0.18 |
ENST00000397022.3
ENST00000353147.5 ENST00000352392.4 ENST00000349999.4 ENST00000317896.7 ENST00000357618.5 ENST00000470149.1 ENST00000336625.8 ENST00000461397.1 ENST00000488199.1 |
HFE
|
hemochromatosis |
| chr4_-_103748271 | 0.18 |
ENST00000343106.5
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr6_+_34433844 | 0.18 |
ENST00000244458.2
ENST00000374043.2 |
PACSIN1
|
protein kinase C and casein kinase substrate in neurons 1 |
| chr3_+_105086056 | 0.18 |
ENST00000472644.2
|
ALCAM
|
activated leukocyte cell adhesion molecule |
| chr3_-_135915146 | 0.18 |
ENST00000473093.1
|
MSL2
|
male-specific lethal 2 homolog (Drosophila) |
| chr14_-_23479331 | 0.18 |
ENST00000397377.1
ENST00000397379.3 ENST00000406429.2 ENST00000341470.4 ENST00000555998.1 ENST00000397376.2 ENST00000553675.1 ENST00000553931.1 ENST00000555575.1 ENST00000553958.1 ENST00000555098.1 ENST00000556419.1 ENST00000553606.1 ENST00000299088.6 ENST00000554179.1 ENST00000397382.4 |
C14orf93
|
chromosome 14 open reading frame 93 |
| chr1_-_21978312 | 0.18 |
ENST00000359708.4
ENST00000290101.4 |
RAP1GAP
|
RAP1 GTPase activating protein |
| chr3_-_52479043 | 0.17 |
ENST00000231721.2
ENST00000475739.1 |
SEMA3G
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3G |
| chr4_+_26859300 | 0.17 |
ENST00000494628.2
|
STIM2
|
stromal interaction molecule 2 |
| chr16_+_53164956 | 0.17 |
ENST00000563410.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr5_+_127419449 | 0.17 |
ENST00000262461.2
ENST00000343225.4 |
SLC12A2
|
solute carrier family 12 (sodium/potassium/chloride transporter), member 2 |
| chr19_+_54041534 | 0.17 |
ENST00000509047.1
ENST00000509585.1 |
ZNF331
|
zinc finger protein 331 |
| chr10_-_135150367 | 0.17 |
ENST00000368555.3
ENST00000252939.4 ENST00000368558.1 ENST00000368556.2 |
CALY
|
calcyon neuron-specific vesicular protein |
| chr6_-_34524049 | 0.17 |
ENST00000374037.3
|
SPDEF
|
SAM pointed domain containing ETS transcription factor |
| chr18_+_74240756 | 0.17 |
ENST00000584910.1
ENST00000582452.1 |
LINC00908
|
long intergenic non-protein coding RNA 908 |
| chr10_+_98592009 | 0.16 |
ENST00000540664.1
ENST00000371103.3 |
LCOR
|
ligand dependent nuclear receptor corepressor |
| chrX_-_30327495 | 0.16 |
ENST00000453287.1
|
NR0B1
|
nuclear receptor subfamily 0, group B, member 1 |
| chr15_-_72612470 | 0.16 |
ENST00000287202.5
|
CELF6
|
CUGBP, Elav-like family member 6 |
| chr12_+_124457746 | 0.16 |
ENST00000392404.3
ENST00000538932.2 ENST00000337815.4 ENST00000540762.2 |
ZNF664
FAM101A
|
zinc finger protein 664 family with sequence similarity 101, member A |
| chr15_+_96873921 | 0.16 |
ENST00000394166.3
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr19_+_49977818 | 0.16 |
ENST00000594009.1
ENST00000595510.1 |
FLT3LG
|
fms-related tyrosine kinase 3 ligand |
| chr2_+_30670077 | 0.16 |
ENST00000466477.1
ENST00000465200.1 ENST00000379509.3 ENST00000319406.4 ENST00000488144.1 ENST00000465538.1 ENST00000309052.4 ENST00000359433.1 |
LCLAT1
|
lysocardiolipin acyltransferase 1 |
| chr2_+_232575168 | 0.16 |
ENST00000440384.1
|
PTMA
|
prothymosin, alpha |
| chr6_-_138428613 | 0.15 |
ENST00000421351.3
|
PERP
|
PERP, TP53 apoptosis effector |
| chr7_+_12250833 | 0.15 |
ENST00000396668.3
|
TMEM106B
|
transmembrane protein 106B |
| chr6_-_34524093 | 0.15 |
ENST00000544425.1
|
SPDEF
|
SAM pointed domain containing ETS transcription factor |
| chr6_-_105584560 | 0.15 |
ENST00000336775.5
|
BVES
|
blood vessel epicardial substance |
| chr5_+_421030 | 0.15 |
ENST00000506456.1
|
AHRR
|
aryl-hydrocarbon receptor repressor |
| chr12_+_69753448 | 0.15 |
ENST00000247843.2
ENST00000548020.1 ENST00000549685.1 ENST00000552955.1 |
YEATS4
|
YEATS domain containing 4 |
| chr19_-_49140609 | 0.15 |
ENST00000601104.1
|
DBP
|
D site of albumin promoter (albumin D-box) binding protein |
| chr3_+_16926441 | 0.15 |
ENST00000418129.2
ENST00000396755.2 |
PLCL2
|
phospholipase C-like 2 |
| chr2_-_219433014 | 0.15 |
ENST00000418019.1
ENST00000454775.1 ENST00000338465.5 ENST00000415516.1 ENST00000258399.3 |
USP37
|
ubiquitin specific peptidase 37 |
| chr5_+_6633456 | 0.15 |
ENST00000274192.5
|
SRD5A1
|
steroid-5-alpha-reductase, alpha polypeptide 1 (3-oxo-5 alpha-steroid delta 4-dehydrogenase alpha 1) |
| chr4_+_187065978 | 0.14 |
ENST00000227065.4
ENST00000502970.1 ENST00000514153.1 |
FAM149A
|
family with sequence similarity 149, member A |
| chr5_-_142782862 | 0.14 |
ENST00000415690.2
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr7_+_12250886 | 0.14 |
ENST00000444443.1
ENST00000396667.3 |
TMEM106B
|
transmembrane protein 106B |
| chr12_-_123717711 | 0.14 |
ENST00000537854.1
|
MPHOSPH9
|
M-phase phosphoprotein 9 |
| chr9_+_139871948 | 0.14 |
ENST00000224167.2
ENST00000457950.1 ENST00000371625.3 ENST00000371623.3 |
PTGDS
|
prostaglandin D2 synthase 21kDa (brain) |
| chr17_+_79679299 | 0.14 |
ENST00000331531.5
|
SLC25A10
|
solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 |
| chr20_+_19738792 | 0.14 |
ENST00000412571.1
|
RP1-122P22.2
|
RP1-122P22.2 |
| chrX_-_107019181 | 0.14 |
ENST00000315660.4
ENST00000372384.2 ENST00000502650.1 ENST00000506724.1 |
TSC22D3
|
TSC22 domain family, member 3 |
| chr17_+_260097 | 0.14 |
ENST00000360127.6
ENST00000571106.1 ENST00000491373.1 |
C17orf97
|
chromosome 17 open reading frame 97 |
| chr20_+_34680595 | 0.14 |
ENST00000406771.2
|
EPB41L1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr17_-_27229948 | 0.14 |
ENST00000426464.2
|
DHRS13
|
dehydrogenase/reductase (SDR family) member 13 |
| chr17_+_4613918 | 0.13 |
ENST00000574954.1
ENST00000346341.2 ENST00000572457.1 ENST00000381488.6 ENST00000412477.3 ENST00000571428.1 ENST00000575877.1 |
ARRB2
|
arrestin, beta 2 |
| chr17_-_73179046 | 0.13 |
ENST00000314523.7
ENST00000420826.2 |
SUMO2
|
small ubiquitin-like modifier 2 |
| chr22_+_21311367 | 0.13 |
ENST00000444039.1
|
XXbac-B135H6.15
|
XXbac-B135H6.15 |
| chr4_+_191001979 | 0.13 |
ENST00000538692.1
|
DUX4L4
|
double homeobox 4 like 4 |
| chr4_-_89080003 | 0.13 |
ENST00000237612.3
|
ABCG2
|
ATP-binding cassette, sub-family G (WHITE), member 2 |
| chr19_-_49568311 | 0.13 |
ENST00000595857.1
ENST00000451356.2 |
NTF4
|
neurotrophin 4 |
| chr12_-_64616019 | 0.13 |
ENST00000311915.8
ENST00000398055.3 ENST00000544871.1 |
C12orf66
|
chromosome 12 open reading frame 66 |
| chrX_-_20284733 | 0.13 |
ENST00000438357.1
|
RPS6KA3
|
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| chr20_-_44519839 | 0.13 |
ENST00000372518.4
|
NEURL2
|
neuralized E3 ubiquitin protein ligase 2 |
| chr3_-_179169181 | 0.13 |
ENST00000497513.1
|
GNB4
|
guanine nucleotide binding protein (G protein), beta polypeptide 4 |
| chr19_+_544034 | 0.13 |
ENST00000592501.1
ENST00000264553.3 |
GZMM
|
granzyme M (lymphocyte met-ase 1) |
| chr5_+_139028510 | 0.13 |
ENST00000502336.1
ENST00000520967.1 ENST00000511048.1 |
CXXC5
|
CXXC finger protein 5 |
| chr12_+_124457670 | 0.13 |
ENST00000539644.1
|
ZNF664
|
zinc finger protein 664 |
| chr1_+_6845578 | 0.13 |
ENST00000467404.2
ENST00000439411.2 |
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr1_-_21948906 | 0.13 |
ENST00000374761.2
ENST00000599760.1 |
RAP1GAP
|
RAP1 GTPase activating protein |
| chr3_-_138048631 | 0.13 |
ENST00000484930.1
ENST00000475751.1 |
NME9
|
NME/NM23 family member 9 |
| chr3_+_105085734 | 0.13 |
ENST00000306107.5
|
ALCAM
|
activated leukocyte cell adhesion molecule |
| chr7_-_72972319 | 0.13 |
ENST00000223368.2
|
BCL7B
|
B-cell CLL/lymphoma 7B |
| chr16_+_21312170 | 0.13 |
ENST00000338573.5
ENST00000561968.1 |
CRYM-AS1
|
CRYM antisense RNA 1 |
| chr15_-_45670924 | 0.13 |
ENST00000396659.3
|
GATM
|
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr1_+_48688357 | 0.13 |
ENST00000533824.1
ENST00000438567.2 ENST00000236495.5 ENST00000420136.2 |
SLC5A9
|
solute carrier family 5 (sodium/sugar cotransporter), member 9 |
| chr19_-_6110555 | 0.13 |
ENST00000593241.1
|
RFX2
|
regulatory factor X, 2 (influences HLA class II expression) |
| chr16_-_4166186 | 0.12 |
ENST00000294016.3
|
ADCY9
|
adenylate cyclase 9 |
| chr8_+_110552046 | 0.12 |
ENST00000529931.1
|
EBAG9
|
estrogen receptor binding site associated, antigen, 9 |
| chr22_-_42322795 | 0.12 |
ENST00000291232.3
|
TNFRSF13C
|
tumor necrosis factor receptor superfamily, member 13C |
| chr7_-_72971934 | 0.12 |
ENST00000411832.1
|
BCL7B
|
B-cell CLL/lymphoma 7B |
| chr11_+_61447845 | 0.12 |
ENST00000257215.5
|
DAGLA
|
diacylglycerol lipase, alpha |
| chr18_+_42260861 | 0.12 |
ENST00000282030.5
|
SETBP1
|
SET binding protein 1 |
| chr5_-_142783694 | 0.12 |
ENST00000394466.2
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr11_-_86383157 | 0.12 |
ENST00000393324.3
|
ME3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial |
| chr10_+_103911926 | 0.12 |
ENST00000605788.1
ENST00000405356.1 |
NOLC1
|
nucleolar and coiled-body phosphoprotein 1 |
| chr2_+_26785409 | 0.12 |
ENST00000329615.3
ENST00000409392.1 |
C2orf70
|
chromosome 2 open reading frame 70 |
| chr1_+_227127981 | 0.12 |
ENST00000366778.1
ENST00000366777.3 ENST00000458507.2 |
ADCK3
|
aarF domain containing kinase 3 |
| chr5_-_159739532 | 0.12 |
ENST00000520748.1
ENST00000393977.3 ENST00000257536.7 |
CCNJL
|
cyclin J-like |
| chr3_-_44552094 | 0.12 |
ENST00000436261.1
|
ZNF852
|
zinc finger protein 852 |
| chr19_+_54058073 | 0.12 |
ENST00000505949.1
ENST00000513265.1 |
ZNF331
|
zinc finger protein 331 |
| chr16_-_70719925 | 0.12 |
ENST00000338779.6
|
MTSS1L
|
metastasis suppressor 1-like |
| chr12_-_88974236 | 0.12 |
ENST00000228280.5
ENST00000552044.1 ENST00000357116.4 |
KITLG
|
KIT ligand |
| chr3_+_52570610 | 0.12 |
ENST00000307106.3
ENST00000477703.1 ENST00000476842.1 |
SMIM4
|
small integral membrane protein 4 |
| chr17_+_79679369 | 0.12 |
ENST00000350690.5
|
SLC25A10
|
solute carrier family 25 (mitochondrial carrier; dicarboxylate transporter), member 10 |
| chr3_+_49044765 | 0.12 |
ENST00000429900.2
|
WDR6
|
WD repeat domain 6 |
| chr17_+_59489112 | 0.12 |
ENST00000335108.2
|
C17orf82
|
chromosome 17 open reading frame 82 |
| chr6_+_43543864 | 0.12 |
ENST00000372236.4
ENST00000535400.1 |
POLH
|
polymerase (DNA directed), eta |
| chr11_-_86383370 | 0.12 |
ENST00000526834.1
ENST00000359636.2 |
ME3
|
malic enzyme 3, NADP(+)-dependent, mitochondrial |
| chr4_+_190992087 | 0.11 |
ENST00000553598.1
|
DUX4L7
|
double homeobox 4 like 7 |
| chr10_+_88718314 | 0.11 |
ENST00000348795.4
|
SNCG
|
synuclein, gamma (breast cancer-specific protein 1) |
| chr6_-_8435706 | 0.11 |
ENST00000379660.4
|
SLC35B3
|
solute carrier family 35 (adenosine 3'-phospho 5'-phosphosulfate transporter), member B3 |
| chr10_-_124768300 | 0.11 |
ENST00000368886.5
|
IKZF5
|
IKAROS family zinc finger 5 (Pegasus) |
| chr18_-_74534232 | 0.11 |
ENST00000585258.1
|
RP11-162A12.2
|
Uncharacterized protein |
| chr3_+_153839149 | 0.11 |
ENST00000465093.1
ENST00000465817.1 |
ARHGEF26
|
Rho guanine nucleotide exchange factor (GEF) 26 |
| chr17_-_28257080 | 0.11 |
ENST00000579954.1
ENST00000540801.1 ENST00000269033.3 ENST00000590153.1 ENST00000582084.1 |
SSH2
|
slingshot protein phosphatase 2 |
| chr4_+_6642404 | 0.11 |
ENST00000507420.1
ENST00000382581.4 |
MRFAP1
|
Morf4 family associated protein 1 |
| chr1_-_6295975 | 0.11 |
ENST00000343813.5
ENST00000362035.3 |
ICMT
|
isoprenylcysteine carboxyl methyltransferase |
| chr1_+_6105974 | 0.11 |
ENST00000378083.3
|
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
| chr17_+_76311791 | 0.11 |
ENST00000586321.1
|
AC061992.2
|
AC061992.2 |
| chr15_-_65117807 | 0.11 |
ENST00000559239.1
ENST00000268043.4 ENST00000333425.6 |
PIF1
|
PIF1 5'-to-3' DNA helicase |
| chr17_+_4613776 | 0.11 |
ENST00000269260.2
|
ARRB2
|
arrestin, beta 2 |
| chr4_-_104119528 | 0.11 |
ENST00000380026.3
ENST00000503705.1 ENST00000265148.3 |
CENPE
|
centromere protein E, 312kDa |
| chr2_-_264024 | 0.11 |
ENST00000403712.2
ENST00000356150.5 ENST00000405430.1 |
SH3YL1
|
SH3 and SYLF domain containing 1 |
| chr1_+_161068179 | 0.11 |
ENST00000368011.4
ENST00000392192.2 |
KLHDC9
|
kelch domain containing 9 |
| chr9_-_14322319 | 0.11 |
ENST00000606230.1
|
NFIB
|
nuclear factor I/B |
| chr1_+_1407133 | 0.11 |
ENST00000378741.3
ENST00000308647.7 |
ATAD3B
|
ATPase family, AAA domain containing 3B |
| chr2_-_128615681 | 0.11 |
ENST00000409955.1
ENST00000272645.4 |
POLR2D
|
polymerase (RNA) II (DNA directed) polypeptide D |
| chr12_+_82752283 | 0.11 |
ENST00000548200.1
|
METTL25
|
methyltransferase like 25 |
| chr8_-_125740514 | 0.11 |
ENST00000325064.5
ENST00000518547.1 |
MTSS1
|
metastasis suppressor 1 |
| chr1_+_234509413 | 0.11 |
ENST00000366613.1
ENST00000366612.1 |
COA6
|
cytochrome c oxidase assembly factor 6 homolog (S. cerevisiae) |
| chr9_-_135545380 | 0.11 |
ENST00000544003.1
|
DDX31
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 31 |
| chr10_-_25012597 | 0.11 |
ENST00000396432.2
|
ARHGAP21
|
Rho GTPase activating protein 21 |
| chr19_+_41036371 | 0.11 |
ENST00000392023.1
|
SPTBN4
|
spectrin, beta, non-erythrocytic 4 |
| chr6_+_26087646 | 0.11 |
ENST00000309234.6
|
HFE
|
hemochromatosis |
| chr19_+_3359561 | 0.11 |
ENST00000589123.1
ENST00000346156.5 ENST00000395111.3 ENST00000586919.1 |
NFIC
|
nuclear factor I/C (CCAAT-binding transcription factor) |
| chr4_-_76598326 | 0.11 |
ENST00000503660.1
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr2_+_20646824 | 0.11 |
ENST00000272233.4
|
RHOB
|
ras homolog family member B |
| chr11_-_27384737 | 0.11 |
ENST00000317945.6
|
CCDC34
|
coiled-coil domain containing 34 |
| chr9_-_113018746 | 0.11 |
ENST00000374515.5
|
TXN
|
thioredoxin |
| chr12_+_123849462 | 0.11 |
ENST00000543072.1
|
hsa-mir-8072
|
hsa-mir-8072 |
| chr4_+_15004165 | 0.11 |
ENST00000538197.1
ENST00000541112.1 ENST00000442003.2 |
CPEB2
|
cytoplasmic polyadenylation element binding protein 2 |
| chr15_-_57025759 | 0.11 |
ENST00000267807.7
|
ZNF280D
|
zinc finger protein 280D |
| chr17_+_76210267 | 0.11 |
ENST00000301633.4
ENST00000350051.3 ENST00000374948.2 ENST00000590449.1 |
BIRC5
|
baculoviral IAP repeat containing 5 |
| chr19_+_32896646 | 0.11 |
ENST00000392250.2
|
DPY19L3
|
dpy-19-like 3 (C. elegans) |
| chrX_-_63425561 | 0.11 |
ENST00000374869.3
ENST00000330258.3 |
AMER1
|
APC membrane recruitment protein 1 |
| chr20_-_61733657 | 0.11 |
ENST00000608031.1
ENST00000447910.2 |
HAR1B
|
highly accelerated region 1B (non-protein coding) |
| chr1_+_245027971 | 0.11 |
ENST00000610145.1
|
RP11-11N7.4
|
RP11-11N7.4 |
| chr8_-_125740730 | 0.11 |
ENST00000354184.4
|
MTSS1
|
metastasis suppressor 1 |
| chr19_+_54041333 | 0.11 |
ENST00000411977.2
ENST00000511154.1 |
ZNF331
|
zinc finger protein 331 |
| chr22_+_45809560 | 0.10 |
ENST00000342894.3
ENST00000538017.1 |
RIBC2
|
RIB43A domain with coiled-coils 2 |
| chr14_-_100070363 | 0.10 |
ENST00000380243.4
|
CCDC85C
|
coiled-coil domain containing 85C |
| chr10_+_134210672 | 0.10 |
ENST00000305233.5
ENST00000368609.4 |
PWWP2B
|
PWWP domain containing 2B |
| chr2_+_231902193 | 0.10 |
ENST00000373640.4
|
C2orf72
|
chromosome 2 open reading frame 72 |
| chr6_+_170102210 | 0.10 |
ENST00000439249.1
ENST00000332290.2 |
C6orf120
|
chromosome 6 open reading frame 120 |
| chr20_+_18118703 | 0.10 |
ENST00000464792.1
|
CSRP2BP
|
CSRP2 binding protein |
| chr8_-_144679264 | 0.10 |
ENST00000531953.1
ENST00000526133.1 |
EEF1D
|
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chr7_-_27169801 | 0.10 |
ENST00000511914.1
|
HOXA4
|
homeobox A4 |
| chr2_-_27341966 | 0.10 |
ENST00000402394.1
ENST00000402550.1 ENST00000260595.5 |
CGREF1
|
cell growth regulator with EF-hand domain 1 |
| chr3_-_197282821 | 0.10 |
ENST00000445160.2
ENST00000446746.1 ENST00000432819.1 ENST00000392379.1 ENST00000441275.1 ENST00000392378.2 |
BDH1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr1_+_203274639 | 0.10 |
ENST00000290551.4
|
BTG2
|
BTG family, member 2 |
| chr5_-_130970723 | 0.10 |
ENST00000308008.6
ENST00000296859.6 ENST00000507093.1 ENST00000510071.1 ENST00000509018.1 ENST00000307984.5 |
RAPGEF6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr20_+_36531499 | 0.10 |
ENST00000373458.3
ENST00000373461.4 ENST00000373459.4 |
VSTM2L
|
V-set and transmembrane domain containing 2 like |
| chr10_-_43133950 | 0.10 |
ENST00000359467.3
|
ZNF33B
|
zinc finger protein 33B |
| chr4_+_106816592 | 0.10 |
ENST00000379987.2
ENST00000453617.2 ENST00000427316.2 ENST00000514622.1 ENST00000305572.8 |
NPNT
|
nephronectin |
| chr14_-_24780573 | 0.10 |
ENST00000336557.5
ENST00000258807.5 |
CIDEB
|
cell death-inducing DFFA-like effector b |
| chr9_-_100000957 | 0.10 |
ENST00000366109.2
ENST00000607322.1 |
RP11-498P14.5
|
RP11-498P14.5 |
| chr8_+_85095553 | 0.10 |
ENST00000521268.1
|
RALYL
|
RALY RNA binding protein-like |
| chr2_-_107503558 | 0.10 |
ENST00000361686.4
ENST00000409087.3 |
ST6GAL2
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 2 |
| chr11_+_102980251 | 0.10 |
ENST00000334267.7
ENST00000398093.3 |
DYNC2H1
|
dynein, cytoplasmic 2, heavy chain 1 |
| chr7_-_21985489 | 0.10 |
ENST00000356195.5
ENST00000447180.1 ENST00000373934.4 ENST00000457951.1 |
CDCA7L
|
cell division cycle associated 7-like |
| chr5_-_176730733 | 0.10 |
ENST00000504395.1
|
RAB24
|
RAB24, member RAS oncogene family |
| chr10_+_64564469 | 0.10 |
ENST00000373783.1
|
ADO
|
2-aminoethanethiol (cysteamine) dioxygenase |
| chr1_+_6845497 | 0.10 |
ENST00000473578.1
ENST00000557126.1 |
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr5_-_180287663 | 0.10 |
ENST00000509066.1
ENST00000504225.1 |
ZFP62
|
ZFP62 zinc finger protein |
| chr16_-_87525651 | 0.10 |
ENST00000268616.4
|
ZCCHC14
|
zinc finger, CCHC domain containing 14 |
| chr6_-_105585022 | 0.10 |
ENST00000314641.5
|
BVES
|
blood vessel epicardial substance |
| chr19_+_10362882 | 0.10 |
ENST00000393733.2
ENST00000588502.1 |
MRPL4
|
mitochondrial ribosomal protein L4 |
| chr5_-_10308125 | 0.10 |
ENST00000296658.3
|
CMBL
|
carboxymethylenebutenolidase homolog (Pseudomonas) |
| chr6_-_10747802 | 0.10 |
ENST00000606522.1
ENST00000606652.1 |
RP11-421M1.8
|
RP11-421M1.8 |
| chr3_+_157261116 | 0.10 |
ENST00000468043.1
ENST00000459838.1 ENST00000461040.1 ENST00000449199.2 ENST00000426338.2 |
C3orf55
|
chromosome 3 open reading frame 55 |
| chr16_+_777118 | 0.10 |
ENST00000562141.1
|
HAGHL
|
hydroxyacylglutathione hydrolase-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of AHR_ARNT2
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:1990709 | presynaptic active zone organization(GO:1990709) |
| 0.2 | 1.1 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.1 | 0.3 | GO:1900390 | positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) |
| 0.1 | 0.3 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 0.1 | 0.3 | GO:0015709 | thiosulfate transport(GO:0015709) oxaloacetate transport(GO:0015729) malate transport(GO:0015743) malate transmembrane transport(GO:0071423) oxaloacetate(2-) transmembrane transport(GO:1902356) |
| 0.1 | 0.2 | GO:0060931 | sinoatrial node cell development(GO:0060931) |
| 0.1 | 0.5 | GO:2000490 | negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.1 | 0.2 | GO:1903489 | epithelial cell differentiation involved in salivary gland development(GO:0060690) epithelial cell maturation involved in salivary gland development(GO:0060691) regulation of plasma cell differentiation(GO:1900098) positive regulation of plasma cell differentiation(GO:1900100) regulation of lactation(GO:1903487) positive regulation of lactation(GO:1903489) |
| 0.1 | 0.3 | GO:0006601 | creatine biosynthetic process(GO:0006601) |
| 0.1 | 0.2 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.1 | 0.2 | GO:0035623 | renal glucose absorption(GO:0035623) |
| 0.1 | 0.2 | GO:0034402 | recruitment of 3'-end processing factors to RNA polymerase II holoenzyme complex(GO:0034402) |
| 0.1 | 0.3 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 0.6 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.2 | GO:0030885 | regulation of myeloid dendritic cell activation(GO:0030885) |
| 0.0 | 0.1 | GO:1902559 | 3'-phosphoadenosine 5'-phosphosulfate transport(GO:0046963) 3'-phospho-5'-adenylyl sulfate transmembrane transport(GO:1902559) |
| 0.0 | 0.1 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.4 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.0 | 0.2 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.2 | GO:1903613 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.0 | 0.3 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.0 | 0.1 | GO:0070446 | cellular response to caloric restriction(GO:0061433) negative regulation of oligodendrocyte progenitor proliferation(GO:0070446) |
| 0.0 | 0.3 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.4 | GO:0071926 | endocannabinoid signaling pathway(GO:0071926) |
| 0.0 | 0.1 | GO:0008052 | sensory organ boundary specification(GO:0008052) formation of organ boundary(GO:0010160) taste bud development(GO:0061193) |
| 0.0 | 0.6 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.2 | GO:0070541 | response to platinum ion(GO:0070541) |
| 0.0 | 0.3 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.0 | GO:1901162 | primary amino compound biosynthetic process(GO:1901162) |
| 0.0 | 0.2 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.0 | 0.1 | GO:0033024 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.0 | 0.2 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.2 | GO:0021623 | optic cup structural organization(GO:0003409) oculomotor nerve morphogenesis(GO:0021622) oculomotor nerve formation(GO:0021623) |
| 0.0 | 0.1 | GO:2000437 | monocyte extravasation(GO:0035696) regulation of monocyte extravasation(GO:2000437) |
| 0.0 | 0.1 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.0 | 0.2 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.1 | GO:0097195 | pilomotor reflex(GO:0097195) |
| 0.0 | 0.1 | GO:0000921 | septin ring assembly(GO:0000921) septin ring organization(GO:0031106) |
| 0.0 | 0.4 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.2 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.5 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.0 | GO:0060086 | circadian temperature homeostasis(GO:0060086) |
| 0.0 | 0.2 | GO:0090272 | negative regulation of fibroblast growth factor production(GO:0090272) |
| 0.0 | 0.3 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.1 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.1 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.0 | 0.3 | GO:0018211 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.0 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.0 | GO:0097069 | cellular response to thyroxine stimulus(GO:0097069) cellular response to L-phenylalanine derivative(GO:1904387) |
| 0.0 | 0.1 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.0 | 0.1 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.0 | 0.1 | GO:1900247 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.0 | 0.1 | GO:1901079 | positive regulation of relaxation of muscle(GO:1901079) |
| 0.0 | 0.1 | GO:0006749 | glutathione metabolic process(GO:0006749) |
| 0.0 | 0.1 | GO:0007057 | spindle assembly involved in female meiosis I(GO:0007057) |
| 0.0 | 0.1 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.0 | 0.1 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.0 | 0.1 | GO:0006433 | prolyl-tRNA aminoacylation(GO:0006433) |
| 0.0 | 0.1 | GO:0003169 | coronary vein morphogenesis(GO:0003169) |
| 0.0 | 0.1 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) |
| 0.0 | 0.1 | GO:0043096 | purine nucleobase salvage(GO:0043096) |
| 0.0 | 0.1 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.0 | 0.1 | GO:2000705 | dense core granule biogenesis(GO:0061110) regulation of dense core granule biogenesis(GO:2000705) |
| 0.0 | 0.2 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 | 0.0 | GO:0031291 | Ran protein signal transduction(GO:0031291) |
| 0.0 | 0.2 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 | 0.1 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.0 | 0.1 | GO:0019285 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.0 | 0.1 | GO:0006203 | dGTP catabolic process(GO:0006203) |
| 0.0 | 0.2 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.0 | GO:0035411 | catenin import into nucleus(GO:0035411) |
| 0.0 | 0.1 | GO:0006880 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.1 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.0 | 0.1 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.0 | 0.2 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.0 | 0.2 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.1 | GO:0051300 | spindle pole body duplication(GO:0030474) spindle pole body organization(GO:0051300) spindle pole body localization(GO:0070631) establishment of spindle pole body localization(GO:0070632) spindle pole body localization to nuclear envelope(GO:0071789) establishment of spindle pole body localization to nuclear envelope(GO:0071790) |
| 0.0 | 0.0 | GO:0036023 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.0 | 0.1 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.0 | 0.3 | GO:0042428 | serotonin metabolic process(GO:0042428) |
| 0.0 | 0.1 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.1 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.0 | 0.1 | GO:0090341 | negative regulation of secretion of lysosomal enzymes(GO:0090341) |
| 0.0 | 0.1 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.1 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 0.1 | GO:1902990 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.0 | 0.1 | GO:2001279 | regulation of prostaglandin biosynthetic process(GO:0031392) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) |
| 0.0 | 0.1 | GO:1901837 | negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.0 | 0.0 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.0 | 0.2 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 0.1 | GO:0010638 | positive regulation of organelle organization(GO:0010638) |
| 0.0 | 0.1 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.0 | 0.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.1 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.1 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) RNA polymerase I transcriptional preinitiation complex assembly(GO:0001188) RNA polymerase I transcriptional preinitiation complex assembly at the promoter for the nuclear large rRNA transcript(GO:0001189) |
| 0.0 | 0.1 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.0 | 0.1 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.0 | 0.2 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.0 | 0.2 | GO:0018916 | nitrobenzene metabolic process(GO:0018916) |
| 0.0 | 0.1 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.0 | 0.1 | GO:0007388 | anterior compartment pattern formation(GO:0007387) posterior compartment specification(GO:0007388) |
| 0.0 | 0.1 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.0 | 0.1 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.1 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.2 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.0 | 0.1 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.0 | 0.1 | GO:0009181 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.0 | 0.0 | GO:0002554 | serotonin production involved in inflammatory response(GO:0002351) serotonin secretion involved in inflammatory response(GO:0002442) serotonin secretion by platelet(GO:0002554) |
| 0.0 | 0.2 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.0 | 0.1 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.1 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.0 | 0.0 | GO:0009996 | negative regulation of cell fate specification(GO:0009996) |
| 0.0 | 0.1 | GO:1902463 | protein localization to cell leading edge(GO:1902463) |
| 0.0 | 0.1 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.0 | 0.0 | GO:0030860 | regulation of polarized epithelial cell differentiation(GO:0030860) |
| 0.0 | 0.1 | GO:0060827 | regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060827) negative regulation of canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060829) |
| 0.0 | 0.3 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.1 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.0 | 0.0 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.0 | GO:0090381 | negative regulation of mesodermal cell fate specification(GO:0042662) regulation of heart induction(GO:0090381) regulation of heart induction by canonical Wnt signaling pathway(GO:0100012) |
| 0.0 | 0.0 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.0 | 0.1 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.0 | 0.0 | GO:0009880 | embryonic pattern specification(GO:0009880) |
| 0.0 | 0.1 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.0 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.0 | 0.0 | GO:0033076 | isoquinoline alkaloid metabolic process(GO:0033076) |
| 0.0 | 0.0 | GO:0002605 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) negative regulation of dendritic cell antigen processing and presentation(GO:0002605) |
| 0.0 | 0.1 | GO:0070982 | L-asparagine biosynthetic process(GO:0070981) L-asparagine metabolic process(GO:0070982) |
| 0.0 | 0.1 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.0 | 0.0 | GO:0006286 | base-excision repair, base-free sugar-phosphate removal(GO:0006286) |
| 0.0 | 0.1 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.0 | 0.1 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.0 | 0.0 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 0.1 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.0 | 0.0 | GO:1990542 | mitochondrial transmembrane transport(GO:1990542) |
| 0.0 | 0.0 | GO:0048243 | norepinephrine secretion(GO:0048243) |
| 0.0 | 0.1 | GO:0042816 | vitamin B6 metabolic process(GO:0042816) |
| 0.0 | 0.0 | GO:0033488 | cholesterol biosynthetic process via 24,25-dihydrolanosterol(GO:0033488) |
| 0.0 | 0.0 | GO:1903452 | regulation of G1 to G0 transition(GO:1903450) positive regulation of G1 to G0 transition(GO:1903452) |
| 0.0 | 0.0 | GO:2000272 | negative regulation of receptor activity(GO:2000272) |
| 0.0 | 0.2 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.0 | 0.2 | GO:0045955 | negative regulation of calcium ion-dependent exocytosis(GO:0045955) |
| 0.0 | 0.0 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.1 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.0 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.0 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.1 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.0 | 0.1 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.1 | GO:1902499 | positive regulation of protein autoubiquitination(GO:1902499) |
| 0.0 | 0.0 | GO:0021557 | oculomotor nerve development(GO:0021557) |
| 0.0 | 0.1 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.0 | 0.0 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.1 | GO:0032902 | nerve growth factor production(GO:0032902) |
| 0.0 | 0.0 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.0 | 0.1 | GO:0001835 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.0 | GO:0061739 | protein lipidation involved in autophagosome assembly(GO:0061739) |
| 0.0 | 0.1 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.0 | 0.0 | GO:0046022 | positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.0 | 0.0 | GO:1904869 | regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) |
| 0.0 | 0.1 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.0 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.0 | 0.0 | GO:2000569 | T-helper 2 cell activation(GO:0035712) regulation of T-helper 2 cell activation(GO:2000569) positive regulation of T-helper 2 cell activation(GO:2000570) |
| 0.0 | 0.0 | GO:0090102 | cochlea development(GO:0090102) |
| 0.0 | 0.0 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.0 | 0.0 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.1 | GO:0031959 | mineralocorticoid receptor signaling pathway(GO:0031959) |
| 0.0 | 0.0 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.0 | GO:0046041 | ITP metabolic process(GO:0046041) |
| 0.0 | 0.0 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.3 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.0 | 0.1 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.0 | 0.0 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.0 | 0.0 | GO:0044837 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.0 | GO:0045799 | positive regulation of chromatin assembly or disassembly(GO:0045799) |
| 0.0 | 0.0 | GO:0003197 | endocardial cushion development(GO:0003197) |
| 0.0 | 0.0 | GO:1902573 | positive regulation of serine-type endopeptidase activity(GO:1900005) positive regulation of serine-type peptidase activity(GO:1902573) |
| 0.0 | 0.0 | GO:0021897 | forebrain astrocyte differentiation(GO:0021896) forebrain astrocyte development(GO:0021897) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.1 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.0 | 0.1 | GO:0032226 | positive regulation of synaptic transmission, dopaminergic(GO:0032226) |
| 0.0 | 0.1 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.0 | 0.0 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.1 | GO:0021764 | amygdala development(GO:0021764) |
| 0.0 | 0.0 | GO:0003407 | neural retina development(GO:0003407) |
| 0.0 | 0.2 | GO:0006183 | GTP biosynthetic process(GO:0006183) |
| 0.0 | 0.0 | GO:0035261 | external genitalia morphogenesis(GO:0035261) |
| 0.0 | 0.0 | GO:1903926 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.0 | 0.1 | GO:1904245 | regulation of polynucleotide adenylyltransferase activity(GO:1904245) |
| 0.0 | 0.0 | GO:0034754 | cellular hormone metabolic process(GO:0034754) |
| 0.0 | 0.1 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.0 | 0.1 | GO:0042797 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 0.1 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.0 | 0.1 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) negative regulation of protein K63-linked ubiquitination(GO:1900045) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.0 | 0.1 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.0 | 0.1 | GO:1900046 | regulation of blood coagulation(GO:0030193) regulation of hemostasis(GO:1900046) |
| 0.0 | 0.1 | GO:1903677 | regulation of cap-independent translational initiation(GO:1903677) positive regulation of cap-independent translational initiation(GO:1903679) regulation of cytoplasmic translational initiation(GO:1904688) positive regulation of cytoplasmic translational initiation(GO:1904690) |
| 0.0 | 0.2 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.1 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.9 | GO:0008542 | visual learning(GO:0008542) |
| 0.0 | 0.1 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.1 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.0 | GO:0032058 | positive regulation of translational initiation in response to stress(GO:0032058) regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) |
| 0.0 | 0.1 | GO:0021987 | cerebral cortex development(GO:0021987) |
| 0.0 | 0.1 | GO:1903845 | negative regulation of cellular response to transforming growth factor beta stimulus(GO:1903845) |
| 0.0 | 0.3 | GO:0044872 | lipoprotein transport(GO:0042953) lipoprotein localization(GO:0044872) |
| 0.0 | 0.0 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.0 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.0 | 0.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.2 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.0 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.0 | 0.0 | GO:0035900 | response to isolation stress(GO:0035900) |
| 0.0 | 0.2 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.0 | GO:1990426 | homologous recombination-dependent replication fork processing(GO:1990426) |
| 0.0 | 0.1 | GO:0010166 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.0 | 0.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.0 | GO:0050993 | dimethylallyl diphosphate biosynthetic process(GO:0050992) dimethylallyl diphosphate metabolic process(GO:0050993) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.1 | 0.3 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.0 | 0.3 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.1 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.2 | GO:0034753 | nuclear aryl hydrocarbon receptor complex(GO:0034753) |
| 0.0 | 0.4 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.3 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.1 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.0 | GO:0000798 | nuclear cohesin complex(GO:0000798) cohesin complex(GO:0008278) |
| 0.0 | 0.1 | GO:0008043 | intracellular ferritin complex(GO:0008043) ferritin complex(GO:0070288) |
| 0.0 | 0.1 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.0 | 0.1 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.1 | GO:0014704 | intercalated disc(GO:0014704) |
| 0.0 | 0.1 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
| 0.0 | 0.3 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.1 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 0.1 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.0 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.1 | GO:1990923 | PET complex(GO:1990923) |
| 0.0 | 0.1 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 0.0 | 0.0 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 0.9 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.1 | GO:0005846 | nuclear cap binding complex(GO:0005846) |
| 0.0 | 0.1 | GO:0097362 | MCM8-MCM9 complex(GO:0097362) |
| 0.0 | 0.1 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 0.0 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.3 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.0 | GO:1903349 | omegasome membrane(GO:1903349) |
| 0.0 | 0.0 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.2 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.1 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.2 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.0 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.1 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.0 | 0.2 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.0 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.3 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.1 | 0.5 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.1 | 0.1 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 0.3 | GO:0015117 | thiosulfate transmembrane transporter activity(GO:0015117) oxaloacetate transmembrane transporter activity(GO:0015131) |
| 0.1 | 0.2 | GO:0031762 | alpha-1A adrenergic receptor binding(GO:0031691) follicle-stimulating hormone receptor binding(GO:0031762) |
| 0.1 | 0.5 | GO:0004473 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.1 | 0.2 | GO:0015068 | amidinotransferase activity(GO:0015067) glycine amidinotransferase activity(GO:0015068) |
| 0.1 | 0.6 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.1 | 0.2 | GO:0004325 | ferrochelatase activity(GO:0004325) |
| 0.1 | 0.2 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.1 | 0.2 | GO:0033765 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 0.1 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 0.1 | GO:1904713 | beta-catenin destruction complex binding(GO:1904713) |
| 0.0 | 0.1 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) phosphotransferase activity, phosphate group as acceptor(GO:0016776) nucleobase-containing compound kinase activity(GO:0019205) |
| 0.0 | 0.1 | GO:0046970 | NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.0 | 0.1 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.0 | 0.1 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.0 | 0.1 | GO:0001181 | transcription factor activity, core RNA polymerase I binding(GO:0001181) |
| 0.0 | 0.1 | GO:0033677 | DNA/RNA helicase activity(GO:0033677) |
| 0.0 | 0.3 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.1 | GO:0035539 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 0.0 | 0.1 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.0 | 0.1 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.0 | 0.1 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.0 | 0.1 | GO:0016418 | S-acetyltransferase activity(GO:0016418) |
| 0.0 | 0.1 | GO:0004119 | cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) |
| 0.0 | 0.0 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.0 | 0.1 | GO:0019961 | interferon binding(GO:0019961) |
| 0.0 | 0.2 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.1 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.0 | 0.0 | GO:0004797 | thymidine kinase activity(GO:0004797) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.0 | 0.1 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.0 | 0.1 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.1 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 0.1 | GO:0004952 | dopamine neurotransmitter receptor activity(GO:0004952) |
| 0.0 | 0.1 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 0.1 | GO:0004827 | proline-tRNA ligase activity(GO:0004827) |
| 0.0 | 0.0 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.1 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.0 | 0.0 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.1 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.0 | 0.1 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 0.1 | GO:0003881 | CDP-diacylglycerol-inositol 3-phosphatidyltransferase activity(GO:0003881) |
| 0.0 | 0.1 | GO:0004796 | thromboxane-A synthase activity(GO:0004796) 12-hydroxyheptadecatrienoic acid synthase activity(GO:0036134) |
| 0.0 | 0.1 | GO:0047017 | prostaglandin-F synthase activity(GO:0047017) |
| 0.0 | 0.1 | GO:0050145 | nucleoside phosphate kinase activity(GO:0050145) |
| 0.0 | 0.1 | GO:0004772 | sterol O-acyltransferase activity(GO:0004772) cholesterol O-acyltransferase activity(GO:0034736) |
| 0.0 | 0.1 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.1 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.1 | GO:0004560 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.0 | 0.1 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) |
| 0.0 | 0.0 | GO:0004315 | 3-oxoacyl-[acyl-carrier-protein] synthase activity(GO:0004315) |
| 0.0 | 0.2 | GO:0004558 | alpha-1,4-glucosidase activity(GO:0004558) maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.6 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.1 | GO:0004974 | leukotriene B4 receptor activity(GO:0001632) leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.1 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.0 | 0.0 | GO:0061711 | N(6)-L-threonylcarbamoyladenine synthase(GO:0061711) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.0 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.0 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.1 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.0 | 0.1 | GO:0050115 | myosin-light-chain-phosphatase activity(GO:0050115) |
| 0.0 | 0.1 | GO:0004074 | biliverdin reductase activity(GO:0004074) |
| 0.0 | 0.0 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.0 | GO:0017130 | poly(C) RNA binding(GO:0017130) |
| 0.0 | 0.0 | GO:0090541 | MIT domain binding(GO:0090541) |
| 0.0 | 0.2 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.0 | GO:0016209 | antioxidant activity(GO:0016209) |
| 0.0 | 0.0 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 0.1 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.0 | 0.1 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.1 | GO:0016979 | lipoate-protein ligase activity(GO:0016979) |
| 0.0 | 0.1 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.0 | 0.0 | GO:0008398 | sterol 14-demethylase activity(GO:0008398) |
| 0.0 | 0.1 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.0 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 0.0 | 0.1 | GO:0031403 | lithium ion binding(GO:0031403) |
| 0.0 | 0.1 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.1 | GO:0004803 | transposase activity(GO:0004803) |
| 0.0 | 0.1 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.0 | 0.1 | GO:0004530 | deoxyribonuclease I activity(GO:0004530) |
| 0.0 | 0.0 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
| 0.0 | 0.0 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.0 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.1 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.1 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.0 | 0.0 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.0 | 0.1 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.2 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.0 | GO:0004057 | arginyltransferase activity(GO:0004057) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 0.0 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.0 | GO:0031862 | prostanoid receptor binding(GO:0031862) |
| 0.0 | 0.0 | GO:0001855 | complement component C4b binding(GO:0001855) |
| 0.0 | 0.1 | GO:0005329 | dopamine transmembrane transporter activity(GO:0005329) |
| 0.0 | 0.0 | GO:0000384 | first spliceosomal transesterification activity(GO:0000384) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.0 | 0.1 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.2 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0001032 | RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.0 | 0.0 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.0 | 0.0 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 0.1 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.0 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.0 | GO:0016165 | linoleate 13S-lipoxygenase activity(GO:0016165) |
| 0.0 | 0.0 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.0 | 0.0 | GO:0035034 | histone acetyltransferase regulator activity(GO:0035034) |
| 0.0 | 0.0 | GO:0019781 | NEDD8 activating enzyme activity(GO:0019781) |
| 0.0 | 0.1 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.0 | GO:0004452 | isopentenyl-diphosphate delta-isomerase activity(GO:0004452) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.0 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.2 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.0 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 1.2 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.4 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.1 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 0.8 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.4 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |