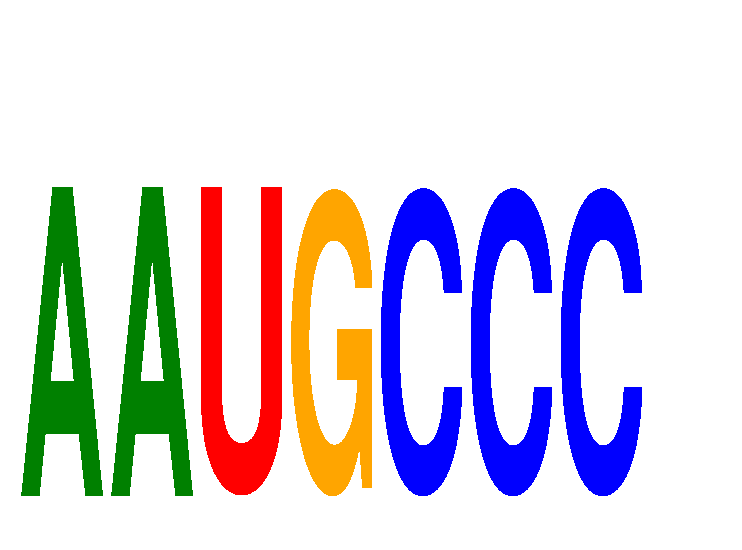Project
A549 cells infected with RSV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for AAUGCCC
Z-value: 0.58
miRNA associated with seed AAUGCCC
| Name | miRBASE accession |
|---|---|
|
hsa-miR-365a-3p
|
MIMAT0000710 |
|
hsa-miR-365b-3p
|
MIMAT0022834 |
Activity profile of AAUGCCC motif
Sorted Z-values of AAUGCCC motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_+_68165657 | 1.44 |
ENST00000243457.3
|
KCNJ2
|
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr2_+_155554797 | 0.56 |
ENST00000295101.2
|
KCNJ3
|
potassium inwardly-rectifying channel, subfamily J, member 3 |
| chr2_-_153574480 | 0.22 |
ENST00000410080.1
|
PRPF40A
|
PRP40 pre-mRNA processing factor 40 homolog A (S. cerevisiae) |
| chr15_+_52121822 | 0.22 |
ENST00000558455.1
ENST00000308580.7 |
TMOD3
|
tropomodulin 3 (ubiquitous) |
| chr3_+_51575596 | 0.18 |
ENST00000409535.2
|
RAD54L2
|
RAD54-like 2 (S. cerevisiae) |
| chr2_+_46926048 | 0.18 |
ENST00000306503.5
|
SOCS5
|
suppressor of cytokine signaling 5 |
| chr2_-_64881018 | 0.16 |
ENST00000313349.3
|
SERTAD2
|
SERTA domain containing 2 |
| chr5_-_39074479 | 0.16 |
ENST00000514735.1
ENST00000296782.5 ENST00000357387.3 |
RICTOR
|
RPTOR independent companion of MTOR, complex 2 |
| chr5_-_142065612 | 0.16 |
ENST00000360966.5
ENST00000411960.1 |
FGF1
|
fibroblast growth factor 1 (acidic) |
| chr8_-_82598511 | 0.15 |
ENST00000449740.2
ENST00000311489.4 ENST00000521360.1 ENST00000519964.1 ENST00000518202.1 |
IMPA1
|
inositol(myo)-1(or 4)-monophosphatase 1 |
| chr3_+_23986748 | 0.15 |
ENST00000312521.4
|
NR1D2
|
nuclear receptor subfamily 1, group D, member 2 |
| chr2_+_61108650 | 0.12 |
ENST00000295025.8
|
REL
|
v-rel avian reticuloendotheliosis viral oncogene homolog |
| chr2_-_200322723 | 0.12 |
ENST00000417098.1
|
SATB2
|
SATB homeobox 2 |
| chr8_+_67579807 | 0.12 |
ENST00000519289.1
ENST00000519561.1 ENST00000521889.1 |
C8orf44-SGK3
C8orf44
|
C8orf44-SGK3 readthrough chromosome 8 open reading frame 44 |
| chr1_-_244013384 | 0.12 |
ENST00000366539.1
|
AKT3
|
v-akt murine thymoma viral oncogene homolog 3 |
| chr5_-_90679145 | 0.12 |
ENST00000265138.3
|
ARRDC3
|
arrestin domain containing 3 |
| chr1_-_78225482 | 0.12 |
ENST00000524778.1
ENST00000370794.3 ENST00000370793.1 ENST00000370792.3 |
USP33
|
ubiquitin specific peptidase 33 |
| chr1_+_2160134 | 0.11 |
ENST00000378536.4
|
SKI
|
v-ski avian sarcoma viral oncogene homolog |
| chr20_+_56884752 | 0.10 |
ENST00000244040.3
|
RAB22A
|
RAB22A, member RAS oncogene family |
| chr2_-_180129484 | 0.10 |
ENST00000428443.3
|
SESTD1
|
SEC14 and spectrin domains 1 |
| chr2_+_179345173 | 0.09 |
ENST00000234453.5
|
PLEKHA3
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 3 |
| chr5_+_172483347 | 0.09 |
ENST00000522692.1
ENST00000296953.2 ENST00000540014.1 ENST00000520420.1 |
CREBRF
|
CREB3 regulatory factor |
| chr1_+_97187318 | 0.09 |
ENST00000609116.1
ENST00000370198.1 ENST00000370197.1 ENST00000426398.2 ENST00000394184.3 |
PTBP2
|
polypyrimidine tract binding protein 2 |
| chr2_-_183903133 | 0.09 |
ENST00000361354.4
|
NCKAP1
|
NCK-associated protein 1 |
| chr1_-_22109682 | 0.09 |
ENST00000400301.1
ENST00000532737.1 |
USP48
|
ubiquitin specific peptidase 48 |
| chr8_-_38239732 | 0.09 |
ENST00000534155.1
ENST00000433384.2 ENST00000317025.8 ENST00000316985.3 |
WHSC1L1
|
Wolf-Hirschhorn syndrome candidate 1-like 1 |
| chr15_+_68346501 | 0.09 |
ENST00000249636.6
|
PIAS1
|
protein inhibitor of activated STAT, 1 |
| chr3_-_123603137 | 0.09 |
ENST00000360304.3
ENST00000359169.1 ENST00000346322.5 ENST00000360772.3 |
MYLK
|
myosin light chain kinase |
| chr4_-_11431389 | 0.09 |
ENST00000002596.5
|
HS3ST1
|
heparan sulfate (glucosamine) 3-O-sulfotransferase 1 |
| chr8_+_124780672 | 0.09 |
ENST00000521166.1
ENST00000334705.7 |
FAM91A1
|
family with sequence similarity 91, member A1 |
| chr7_-_151217001 | 0.08 |
ENST00000262187.5
|
RHEB
|
Ras homolog enriched in brain |
| chr10_-_105615164 | 0.08 |
ENST00000355946.2
ENST00000369774.4 |
SH3PXD2A
|
SH3 and PX domains 2A |
| chr4_-_103748880 | 0.08 |
ENST00000453744.2
ENST00000349311.8 |
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr12_-_75905374 | 0.08 |
ENST00000438169.2
ENST00000229214.4 |
KRR1
|
KRR1, small subunit (SSU) processome component, homolog (yeast) |
| chr3_-_113465065 | 0.08 |
ENST00000497255.1
ENST00000478020.1 ENST00000240922.3 ENST00000493900.1 |
NAA50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr4_+_38665810 | 0.07 |
ENST00000261438.5
ENST00000514033.1 |
KLF3
|
Kruppel-like factor 3 (basic) |
| chr13_-_21476900 | 0.07 |
ENST00000400602.2
ENST00000255305.6 |
XPO4
|
exportin 4 |
| chr10_-_98346801 | 0.07 |
ENST00000371142.4
|
TM9SF3
|
transmembrane 9 superfamily member 3 |
| chr10_-_62149433 | 0.07 |
ENST00000280772.2
|
ANK3
|
ankyrin 3, node of Ranvier (ankyrin G) |
| chr8_+_128426535 | 0.07 |
ENST00000465342.2
|
POU5F1B
|
POU class 5 homeobox 1B |
| chr11_-_57283159 | 0.07 |
ENST00000533263.1
ENST00000278426.3 |
SLC43A1
|
solute carrier family 43 (amino acid system L transporter), member 1 |
| chr3_+_171758344 | 0.06 |
ENST00000336824.4
ENST00000423424.1 |
FNDC3B
|
fibronectin type III domain containing 3B |
| chr12_-_31479045 | 0.06 |
ENST00000539409.1
ENST00000395766.1 |
FAM60A
|
family with sequence similarity 60, member A |
| chr5_-_142783175 | 0.06 |
ENST00000231509.3
ENST00000394464.2 |
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr22_+_32340481 | 0.06 |
ENST00000397492.1
|
YWHAH
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta |
| chr7_+_94285637 | 0.06 |
ENST00000482108.1
ENST00000488574.1 |
PEG10
|
paternally expressed 10 |
| chr1_-_28415204 | 0.06 |
ENST00000373871.3
|
EYA3
|
eyes absent homolog 3 (Drosophila) |
| chr5_-_89825328 | 0.06 |
ENST00000500869.2
ENST00000315948.6 ENST00000509384.1 |
LYSMD3
|
LysM, putative peptidoglycan-binding, domain containing 3 |
| chr10_+_89622870 | 0.06 |
ENST00000371953.3
|
PTEN
|
phosphatase and tensin homolog |
| chrX_-_19988382 | 0.06 |
ENST00000356980.3
ENST00000379687.3 ENST00000379682.4 |
CXorf23
|
chromosome X open reading frame 23 |
| chr11_+_34642656 | 0.05 |
ENST00000257831.3
ENST00000450654.2 |
EHF
|
ets homologous factor |
| chr2_-_157189180 | 0.05 |
ENST00000539077.1
ENST00000424077.1 ENST00000426264.1 ENST00000339562.4 ENST00000421709.1 |
NR4A2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr7_+_139044621 | 0.05 |
ENST00000354926.4
|
C7orf55-LUC7L2
|
C7orf55-LUC7L2 readthrough |
| chr15_+_96873921 | 0.05 |
ENST00000394166.3
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr5_+_122181184 | 0.05 |
ENST00000513881.1
|
SNX24
|
sorting nexin 24 |
| chr18_+_56338618 | 0.05 |
ENST00000348428.3
|
MALT1
|
mucosa associated lymphoid tissue lymphoma translocation gene 1 |
| chr2_-_190649082 | 0.05 |
ENST00000392350.3
ENST00000392349.4 |
ORMDL1
|
ORM1-like 1 (S. cerevisiae) |
| chr8_+_59323823 | 0.05 |
ENST00000399598.2
|
UBXN2B
|
UBX domain protein 2B |
| chr8_+_54764346 | 0.05 |
ENST00000297313.3
ENST00000344277.6 |
RGS20
|
regulator of G-protein signaling 20 |
| chr3_-_48885228 | 0.05 |
ENST00000454963.1
ENST00000296446.8 ENST00000419216.1 ENST00000265563.8 |
PRKAR2A
|
protein kinase, cAMP-dependent, regulatory, type II, alpha |
| chr4_+_41992489 | 0.05 |
ENST00000264451.7
|
SLC30A9
|
solute carrier family 30 (zinc transporter), member 9 |
| chr12_+_72666407 | 0.05 |
ENST00000261180.4
|
TRHDE
|
thyrotropin-releasing hormone degrading enzyme |
| chr3_+_101292939 | 0.05 |
ENST00000265260.3
ENST00000469941.1 ENST00000296024.5 |
PCNP
|
PEST proteolytic signal containing nuclear protein |
| chr3_+_38495333 | 0.05 |
ENST00000352511.4
|
ACVR2B
|
activin A receptor, type IIB |
| chr2_-_39348137 | 0.05 |
ENST00000426016.1
|
SOS1
|
son of sevenless homolog 1 (Drosophila) |
| chr6_+_16129308 | 0.04 |
ENST00000356840.3
ENST00000349606.4 |
MYLIP
|
myosin regulatory light chain interacting protein |
| chr7_+_138145076 | 0.04 |
ENST00000343526.4
|
TRIM24
|
tripartite motif containing 24 |
| chr3_-_79068594 | 0.04 |
ENST00000436010.2
|
ROBO1
|
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr9_-_6007787 | 0.04 |
ENST00000399933.3
ENST00000381461.2 ENST00000513355.2 |
KIAA2026
|
KIAA2026 |
| chr10_+_22610124 | 0.04 |
ENST00000376663.3
|
BMI1
|
BMI1 polycomb ring finger oncogene |
| chr9_+_116638562 | 0.04 |
ENST00000374126.5
ENST00000288466.7 |
ZNF618
|
zinc finger protein 618 |
| chr12_-_67072714 | 0.04 |
ENST00000545666.1
ENST00000398016.3 ENST00000359742.4 ENST00000286445.7 ENST00000538211.1 |
GRIP1
|
glutamate receptor interacting protein 1 |
| chr1_-_91487013 | 0.04 |
ENST00000347275.5
ENST00000370440.1 |
ZNF644
|
zinc finger protein 644 |
| chr15_+_41523335 | 0.04 |
ENST00000334660.5
|
CHP1
|
calcineurin-like EF-hand protein 1 |
| chr10_+_180987 | 0.04 |
ENST00000381591.1
|
ZMYND11
|
zinc finger, MYND-type containing 11 |
| chr1_-_155881156 | 0.04 |
ENST00000539040.1
ENST00000368323.3 |
RIT1
|
Ras-like without CAAX 1 |
| chr5_-_59189545 | 0.04 |
ENST00000340635.6
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr13_+_114238997 | 0.04 |
ENST00000538138.1
ENST00000375370.5 |
TFDP1
|
transcription factor Dp-1 |
| chr1_+_29063271 | 0.04 |
ENST00000373812.3
|
YTHDF2
|
YTH domain family, member 2 |
| chr11_-_31839488 | 0.03 |
ENST00000419022.1
ENST00000379132.3 ENST00000379129.2 |
PAX6
|
paired box 6 |
| chr9_-_136933134 | 0.03 |
ENST00000303407.7
|
BRD3
|
bromodomain containing 3 |
| chr2_+_192542850 | 0.03 |
ENST00000410026.2
|
NABP1
|
nucleic acid binding protein 1 |
| chr12_+_68042495 | 0.03 |
ENST00000344096.3
|
DYRK2
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
| chr1_-_153588334 | 0.03 |
ENST00000476873.1
|
S100A14
|
S100 calcium binding protein A14 |
| chr1_-_35658736 | 0.03 |
ENST00000357214.5
|
SFPQ
|
splicing factor proline/glutamine-rich |
| chr14_-_23526739 | 0.03 |
ENST00000397359.3
ENST00000487137.2 |
CDH24
|
cadherin 24, type 2 |
| chr4_-_74124502 | 0.03 |
ENST00000358602.4
ENST00000330838.6 ENST00000561029.1 |
ANKRD17
|
ankyrin repeat domain 17 |
| chr18_+_43753974 | 0.03 |
ENST00000282059.6
ENST00000321319.6 |
C18orf25
|
chromosome 18 open reading frame 25 |
| chr10_-_120101804 | 0.03 |
ENST00000369183.4
ENST00000369172.4 |
FAM204A
|
family with sequence similarity 204, member A |
| chr3_-_55523966 | 0.03 |
ENST00000474267.1
|
WNT5A
|
wingless-type MMTV integration site family, member 5A |
| chr12_-_15942309 | 0.03 |
ENST00000544064.1
ENST00000543523.1 ENST00000536793.1 |
EPS8
|
epidermal growth factor receptor pathway substrate 8 |
| chr6_-_88411911 | 0.03 |
ENST00000257787.5
|
AKIRIN2
|
akirin 2 |
| chr19_+_33166313 | 0.03 |
ENST00000334176.3
|
RGS9BP
|
regulator of G protein signaling 9 binding protein |
| chr17_-_48072574 | 0.03 |
ENST00000434704.2
|
DLX3
|
distal-less homeobox 3 |
| chr9_+_131445928 | 0.03 |
ENST00000372692.4
|
SET
|
SET nuclear oncogene |
| chr9_-_127905736 | 0.03 |
ENST00000336505.6
ENST00000373549.4 |
SCAI
|
suppressor of cancer cell invasion |
| chr9_+_114659046 | 0.03 |
ENST00000374279.3
|
UGCG
|
UDP-glucose ceramide glucosyltransferase |
| chr7_-_27219632 | 0.03 |
ENST00000470747.4
|
RP1-170O19.20
|
Uncharacterized protein |
| chr17_-_78009647 | 0.03 |
ENST00000310924.2
|
TBC1D16
|
TBC1 domain family, member 16 |
| chr3_-_21792838 | 0.02 |
ENST00000281523.2
|
ZNF385D
|
zinc finger protein 385D |
| chr17_-_27278304 | 0.02 |
ENST00000577226.1
|
PHF12
|
PHD finger protein 12 |
| chr11_-_102714534 | 0.02 |
ENST00000299855.5
|
MMP3
|
matrix metallopeptidase 3 (stromelysin 1, progelatinase) |
| chr11_+_63606373 | 0.02 |
ENST00000402010.2
ENST00000315032.8 ENST00000377809.4 ENST00000413835.2 ENST00000377810.3 |
MARK2
|
MAP/microtubule affinity-regulating kinase 2 |
| chrX_-_153979315 | 0.02 |
ENST00000369575.3
ENST00000369568.4 ENST00000424127.2 |
GAB3
|
GRB2-associated binding protein 3 |
| chr17_+_55333876 | 0.02 |
ENST00000284073.2
|
MSI2
|
musashi RNA-binding protein 2 |
| chr10_+_102505468 | 0.02 |
ENST00000361791.3
ENST00000355243.3 ENST00000428433.1 ENST00000370296.2 |
PAX2
|
paired box 2 |
| chrX_-_3631635 | 0.02 |
ENST00000262848.5
|
PRKX
|
protein kinase, X-linked |
| chr1_+_78470530 | 0.02 |
ENST00000370763.5
|
DNAJB4
|
DnaJ (Hsp40) homolog, subfamily B, member 4 |
| chr13_-_53024725 | 0.02 |
ENST00000378060.4
|
VPS36
|
vacuolar protein sorting 36 homolog (S. cerevisiae) |
| chr21_+_38071430 | 0.02 |
ENST00000290399.6
|
SIM2
|
single-minded family bHLH transcription factor 2 |
| chr17_-_56084578 | 0.02 |
ENST00000582730.2
ENST00000584773.1 ENST00000585096.1 ENST00000258962.4 |
SRSF1
|
serine/arginine-rich splicing factor 1 |
| chr2_-_240322643 | 0.02 |
ENST00000345617.3
|
HDAC4
|
histone deacetylase 4 |
| chr18_-_60987220 | 0.02 |
ENST00000398117.1
|
BCL2
|
B-cell CLL/lymphoma 2 |
| chr12_-_66524482 | 0.02 |
ENST00000446587.2
ENST00000266604.2 |
LLPH
|
LLP homolog, long-term synaptic facilitation (Aplysia) |
| chr2_-_43453734 | 0.02 |
ENST00000282388.3
|
ZFP36L2
|
ZFP36 ring finger protein-like 2 |
| chr8_-_57906362 | 0.02 |
ENST00000262644.4
|
IMPAD1
|
inositol monophosphatase domain containing 1 |
| chr1_+_145611010 | 0.02 |
ENST00000369291.5
|
RNF115
|
ring finger protein 115 |
| chr5_+_148724993 | 0.01 |
ENST00000513661.1
ENST00000329271.3 ENST00000416916.2 |
GRPEL2
|
GrpE-like 2, mitochondrial (E. coli) |
| chr16_-_67840442 | 0.01 |
ENST00000536251.1
ENST00000448631.2 ENST00000602677.1 ENST00000411657.2 ENST00000425512.2 ENST00000317506.3 |
RANBP10
|
RAN binding protein 10 |
| chr3_-_33481835 | 0.01 |
ENST00000283629.3
|
UBP1
|
upstream binding protein 1 (LBP-1a) |
| chr10_-_120514720 | 0.01 |
ENST00000369151.3
ENST00000340214.4 |
CACUL1
|
CDK2-associated, cullin domain 1 |
| chr13_-_46425865 | 0.01 |
ENST00000400405.2
|
SIAH3
|
siah E3 ubiquitin protein ligase family member 3 |
| chr1_+_64239657 | 0.01 |
ENST00000371080.1
ENST00000371079.1 |
ROR1
|
receptor tyrosine kinase-like orphan receptor 1 |
| chr5_-_32444828 | 0.01 |
ENST00000265069.8
|
ZFR
|
zinc finger RNA binding protein |
| chr7_-_141401951 | 0.01 |
ENST00000536163.1
|
KIAA1147
|
KIAA1147 |
| chr10_-_735553 | 0.01 |
ENST00000280886.6
ENST00000423550.1 |
DIP2C
|
DIP2 disco-interacting protein 2 homolog C (Drosophila) |
| chr17_+_28804380 | 0.01 |
ENST00000225724.5
ENST00000451249.2 ENST00000467337.2 ENST00000581721.1 ENST00000414833.2 |
GOSR1
|
golgi SNAP receptor complex member 1 |
| chr21_-_46293586 | 0.01 |
ENST00000445724.2
ENST00000397887.3 |
PTTG1IP
|
pituitary tumor-transforming 1 interacting protein |
| chr14_-_21905395 | 0.01 |
ENST00000430710.3
ENST00000553283.1 |
CHD8
|
chromodomain helicase DNA binding protein 8 |
| chr1_-_46598284 | 0.01 |
ENST00000423209.1
ENST00000262741.5 |
PIK3R3
|
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
| chr22_+_21271714 | 0.01 |
ENST00000354336.3
|
CRKL
|
v-crk avian sarcoma virus CT10 oncogene homolog-like |
| chr11_-_77532050 | 0.01 |
ENST00000308488.6
|
RSF1
|
remodeling and spacing factor 1 |
| chr20_-_39317868 | 0.01 |
ENST00000373313.2
|
MAFB
|
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog B |
| chr7_-_83824169 | 0.01 |
ENST00000265362.4
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr7_-_111846435 | 0.01 |
ENST00000437633.1
ENST00000428084.1 |
DOCK4
|
dedicator of cytokinesis 4 |
| chr2_+_173600671 | 0.00 |
ENST00000409036.1
|
RAPGEF4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr11_+_2466218 | 0.00 |
ENST00000155840.5
|
KCNQ1
|
potassium voltage-gated channel, KQT-like subfamily, member 1 |
| chr7_-_32931387 | 0.00 |
ENST00000304056.4
|
KBTBD2
|
kelch repeat and BTB (POZ) domain containing 2 |
| chr10_-_32636106 | 0.00 |
ENST00000263062.8
ENST00000319778.6 |
EPC1
|
enhancer of polycomb homolog 1 (Drosophila) |
| chr1_+_161284047 | 0.00 |
ENST00000367975.2
ENST00000342751.4 ENST00000432287.2 ENST00000392169.2 ENST00000513009.1 |
SDHC
|
succinate dehydrogenase complex, subunit C, integral membrane protein, 15kDa |
| chr9_+_128024067 | 0.00 |
ENST00000461379.1
ENST00000394084.1 ENST00000394105.2 ENST00000470056.1 ENST00000394104.2 ENST00000265956.4 ENST00000394083.2 ENST00000495955.1 ENST00000467750.1 ENST00000297933.6 |
GAPVD1
|
GTPase activating protein and VPS9 domains 1 |
| chr16_+_67063036 | 0.00 |
ENST00000290858.6
ENST00000564034.1 |
CBFB
|
core-binding factor, beta subunit |
| chr6_+_106546808 | 0.00 |
ENST00000369089.3
|
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr5_-_65017921 | 0.00 |
ENST00000381007.4
|
SGTB
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, beta |
| chr11_-_18656028 | 0.00 |
ENST00000336349.5
|
SPTY2D1
|
SPT2, Suppressor of Ty, domain containing 1 (S. cerevisiae) |
| chr4_-_176923483 | 0.00 |
ENST00000280187.7
ENST00000512509.1 |
GPM6A
|
glycoprotein M6A |
| chr5_+_74632993 | 0.00 |
ENST00000287936.4
|
HMGCR
|
3-hydroxy-3-methylglutaryl-CoA reductase |
Network of associatons between targets according to the STRING database.
First level regulatory network of AAUGCCC
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.4 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.1 | 0.6 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.0 | 0.1 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.0 | 0.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.2 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.2 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.1 | GO:0031064 | negative regulation of histone deacetylation(GO:0031064) |
| 0.0 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.2 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.0 | 0.1 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 0.2 | GO:2000544 | cell chemotaxis to fibroblast growth factor(GO:0035766) endothelial cell chemotaxis to fibroblast growth factor(GO:0035768) regulation of cell chemotaxis to fibroblast growth factor(GO:1904847) regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000544) |
| 0.0 | 0.1 | GO:1903984 | negative regulation of ribosome biogenesis(GO:0090071) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.1 | GO:0072658 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.1 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.0 | 0.1 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.0 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.1 | 1.4 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.0 | 0.1 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.2 | GO:0031403 | lithium ion binding(GO:0031403) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0051800 | phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.0 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |