Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
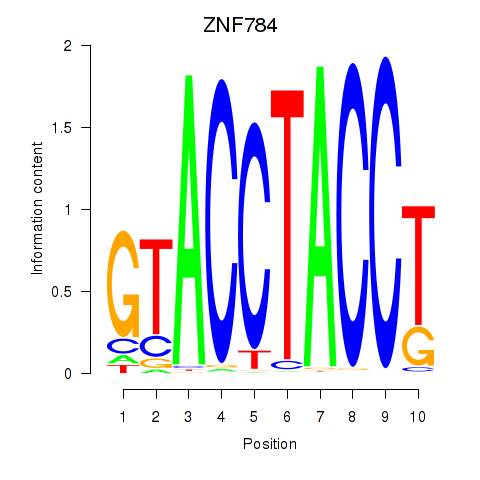
Results for ZNF784
Z-value: 0.60
Transcription factors associated with ZNF784
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF784
|
ENSG00000179922.5 | zinc finger protein 784 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF784 | hg19_v2_chr19_-_56135928_56135967 | -0.84 | 1.6e-01 | Click! |
Activity profile of ZNF784 motif
Sorted Z-values of ZNF784 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_130692631 | 0.23 |
ENST00000500092.2
ENST00000509105.1 |
RP11-519M16.1
|
RP11-519M16.1 |
| chr7_+_26332645 | 0.21 |
ENST00000396376.1
|
SNX10
|
sorting nexin 10 |
| chr8_+_38261880 | 0.20 |
ENST00000527175.1
|
LETM2
|
leucine zipper-EF-hand containing transmembrane protein 2 |
| chr4_-_90758227 | 0.17 |
ENST00000506691.1
ENST00000394986.1 ENST00000506244.1 ENST00000394989.2 ENST00000394991.3 |
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr11_-_119066545 | 0.17 |
ENST00000415318.1
|
CCDC153
|
coiled-coil domain containing 153 |
| chr12_-_12837423 | 0.17 |
ENST00000540510.1
|
GPR19
|
G protein-coupled receptor 19 |
| chr11_+_62652649 | 0.16 |
ENST00000539507.1
|
SLC3A2
|
solute carrier family 3 (amino acid transporter heavy chain), member 2 |
| chr5_+_137225125 | 0.15 |
ENST00000350250.4
ENST00000508638.1 ENST00000502810.1 ENST00000508883.1 |
PKD2L2
|
polycystic kidney disease 2-like 2 |
| chr3_-_47619623 | 0.15 |
ENST00000456150.1
|
CSPG5
|
chondroitin sulfate proteoglycan 5 (neuroglycan C) |
| chr6_-_31550192 | 0.15 |
ENST00000429299.2
ENST00000446745.2 |
LTB
|
lymphotoxin beta (TNF superfamily, member 3) |
| chr20_+_57414795 | 0.14 |
ENST00000371098.2
ENST00000371075.3 |
GNAS
|
GNAS complex locus |
| chr20_+_57414743 | 0.14 |
ENST00000313949.7
|
GNAS
|
GNAS complex locus |
| chr4_+_1340986 | 0.14 |
ENST00000511216.1
ENST00000389851.4 |
UVSSA
|
UV-stimulated scaffold protein A |
| chr1_+_235490659 | 0.13 |
ENST00000488594.1
|
GGPS1
|
geranylgeranyl diphosphate synthase 1 |
| chr3_-_121379739 | 0.13 |
ENST00000428394.2
ENST00000314583.3 |
HCLS1
|
hematopoietic cell-specific Lyn substrate 1 |
| chr1_-_85155939 | 0.13 |
ENST00000603677.1
|
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chr2_-_133104839 | 0.12 |
ENST00000608279.1
|
RP11-725P16.2
|
RP11-725P16.2 |
| chr11_+_45944190 | 0.12 |
ENST00000401752.1
ENST00000389968.3 ENST00000325468.5 ENST00000536139.1 |
GYLTL1B
|
glycosyltransferase-like 1B |
| chr12_+_54402790 | 0.12 |
ENST00000040584.4
|
HOXC8
|
homeobox C8 |
| chr1_-_85156090 | 0.12 |
ENST00000605755.1
ENST00000437941.2 |
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chr5_+_137225158 | 0.12 |
ENST00000290431.5
|
PKD2L2
|
polycystic kidney disease 2-like 2 |
| chr12_-_96389702 | 0.12 |
ENST00000552509.1
|
HAL
|
histidine ammonia-lyase |
| chr6_-_43496605 | 0.11 |
ENST00000455285.2
|
XPO5
|
exportin 5 |
| chrX_+_106045891 | 0.11 |
ENST00000357242.5
ENST00000310452.2 ENST00000481617.2 ENST00000276175.3 |
TBC1D8B
|
TBC1 domain family, member 8B (with GRAM domain) |
| chr13_+_24153488 | 0.11 |
ENST00000382258.4
ENST00000382263.3 |
TNFRSF19
|
tumor necrosis factor receptor superfamily, member 19 |
| chr4_+_48833160 | 0.11 |
ENST00000506801.1
|
OCIAD1
|
OCIA domain containing 1 |
| chr19_-_4540486 | 0.11 |
ENST00000306390.6
|
LRG1
|
leucine-rich alpha-2-glycoprotein 1 |
| chr10_+_93170081 | 0.11 |
ENST00000446394.1
ENST00000371681.4 ENST00000298068.5 |
HECTD2
|
HECT domain containing E3 ubiquitin protein ligase 2 |
| chr1_+_54411995 | 0.11 |
ENST00000319223.4
ENST00000444987.1 |
LRRC42
|
leucine rich repeat containing 42 |
| chr2_+_45878790 | 0.11 |
ENST00000306156.3
|
PRKCE
|
protein kinase C, epsilon |
| chr7_+_22766766 | 0.11 |
ENST00000426291.1
ENST00000401651.1 ENST00000258743.5 ENST00000420258.2 ENST00000407492.1 ENST00000401630.3 ENST00000406575.1 |
IL6
|
interleukin 6 (interferon, beta 2) |
| chr6_-_150039249 | 0.11 |
ENST00000543571.1
|
LATS1
|
large tumor suppressor kinase 1 |
| chr17_-_73149921 | 0.11 |
ENST00000481647.1
ENST00000470924.1 |
HN1
|
hematological and neurological expressed 1 |
| chrX_+_70798261 | 0.11 |
ENST00000373696.3
|
ACRC
|
acidic repeat containing |
| chr1_+_173837488 | 0.11 |
ENST00000427304.1
ENST00000432989.1 ENST00000367702.1 |
ZBTB37
|
zinc finger and BTB domain containing 37 |
| chr11_+_65122216 | 0.10 |
ENST00000309880.5
|
TIGD3
|
tigger transposable element derived 3 |
| chr1_+_90287480 | 0.10 |
ENST00000394593.3
|
LRRC8D
|
leucine rich repeat containing 8 family, member D |
| chr9_-_115480303 | 0.10 |
ENST00000374234.1
ENST00000374238.1 ENST00000374236.1 ENST00000374242.4 |
INIP
|
INTS3 and NABP interacting protein |
| chr4_-_90756769 | 0.10 |
ENST00000345009.4
ENST00000505199.1 ENST00000502987.1 |
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr12_+_100661156 | 0.10 |
ENST00000360820.2
|
SCYL2
|
SCY1-like 2 (S. cerevisiae) |
| chr2_+_170590321 | 0.10 |
ENST00000392647.2
|
KLHL23
|
kelch-like family member 23 |
| chr10_+_92980517 | 0.10 |
ENST00000336126.5
|
PCGF5
|
polycomb group ring finger 5 |
| chr12_-_49525175 | 0.10 |
ENST00000336023.5
ENST00000550367.1 ENST00000552984.1 ENST00000547476.1 |
TUBA1B
|
tubulin, alpha 1b |
| chr18_-_45456930 | 0.09 |
ENST00000262160.6
ENST00000587269.1 |
SMAD2
|
SMAD family member 2 |
| chr21_-_36421626 | 0.09 |
ENST00000300305.3
|
RUNX1
|
runt-related transcription factor 1 |
| chr19_-_376011 | 0.09 |
ENST00000342640.4
|
THEG
|
theg spermatid protein |
| chr4_+_48833234 | 0.09 |
ENST00000510824.1
ENST00000425583.2 |
OCIAD1
|
OCIA domain containing 1 |
| chrX_-_55024967 | 0.09 |
ENST00000545676.1
|
PFKFB1
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1 |
| chr19_+_18284477 | 0.09 |
ENST00000407280.3
|
IFI30
|
interferon, gamma-inducible protein 30 |
| chrX_-_57147748 | 0.09 |
ENST00000374910.3
|
SPIN2B
|
spindlin family, member 2B |
| chr8_+_11561660 | 0.08 |
ENST00000526716.1
ENST00000335135.4 ENST00000528027.1 |
GATA4
|
GATA binding protein 4 |
| chrX_-_135056216 | 0.08 |
ENST00000305963.2
|
MMGT1
|
membrane magnesium transporter 1 |
| chr11_-_31531121 | 0.08 |
ENST00000532287.1
ENST00000526776.1 ENST00000534812.1 ENST00000529749.1 ENST00000278200.1 ENST00000530023.1 ENST00000533642.1 |
IMMP1L
|
IMP1 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr3_+_197476621 | 0.08 |
ENST00000241502.4
|
FYTTD1
|
forty-two-three domain containing 1 |
| chr11_-_64900791 | 0.08 |
ENST00000531018.1
|
SYVN1
|
synovial apoptosis inhibitor 1, synoviolin |
| chr22_+_21321447 | 0.08 |
ENST00000434714.1
|
AIFM3
|
apoptosis-inducing factor, mitochondrion-associated, 3 |
| chr22_-_44708731 | 0.08 |
ENST00000381176.4
|
KIAA1644
|
KIAA1644 |
| chr7_-_558876 | 0.08 |
ENST00000354513.5
ENST00000402802.3 |
PDGFA
|
platelet-derived growth factor alpha polypeptide |
| chr6_-_109804412 | 0.08 |
ENST00000230122.3
|
ZBTB24
|
zinc finger and BTB domain containing 24 |
| chr15_+_89346699 | 0.08 |
ENST00000558207.1
|
ACAN
|
aggrecan |
| chr9_-_110251836 | 0.08 |
ENST00000374672.4
|
KLF4
|
Kruppel-like factor 4 (gut) |
| chr1_-_154155675 | 0.08 |
ENST00000330188.9
ENST00000341485.5 |
TPM3
|
tropomyosin 3 |
| chr17_+_66245341 | 0.08 |
ENST00000577985.1
|
AMZ2
|
archaelysin family metallopeptidase 2 |
| chr20_-_33539655 | 0.08 |
ENST00000451957.2
|
GSS
|
glutathione synthetase |
| chr4_+_48833312 | 0.08 |
ENST00000508293.1
ENST00000513391.2 |
OCIAD1
|
OCIA domain containing 1 |
| chr21_-_36421535 | 0.08 |
ENST00000416754.1
ENST00000437180.1 ENST00000455571.1 |
RUNX1
|
runt-related transcription factor 1 |
| chr7_+_114562172 | 0.08 |
ENST00000393486.1
ENST00000257724.3 |
MDFIC
|
MyoD family inhibitor domain containing |
| chr4_+_48833015 | 0.08 |
ENST00000509122.1
ENST00000509664.1 ENST00000505922.2 ENST00000514981.1 |
OCIAD1
|
OCIA domain containing 1 |
| chr3_+_32993065 | 0.08 |
ENST00000330953.5
|
CCR4
|
chemokine (C-C motif) receptor 4 |
| chr1_-_85156216 | 0.07 |
ENST00000342203.3
ENST00000370612.4 |
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chr8_+_21914477 | 0.07 |
ENST00000517804.1
|
DMTN
|
dematin actin binding protein |
| chr14_+_75894714 | 0.07 |
ENST00000559060.1
|
JDP2
|
Jun dimerization protein 2 |
| chr11_+_60614946 | 0.07 |
ENST00000545580.1
|
CCDC86
|
coiled-coil domain containing 86 |
| chr15_+_48413169 | 0.07 |
ENST00000341459.3
ENST00000482911.2 |
SLC24A5
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 5 |
| chr1_+_228327943 | 0.07 |
ENST00000366726.1
ENST00000312726.4 ENST00000366728.2 ENST00000453943.1 ENST00000366723.1 ENST00000366722.1 ENST00000435153.1 ENST00000366721.1 |
GUK1
|
guanylate kinase 1 |
| chr2_-_145277640 | 0.07 |
ENST00000539609.3
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr9_-_139094988 | 0.07 |
ENST00000371746.3
|
LHX3
|
LIM homeobox 3 |
| chr19_+_13858701 | 0.07 |
ENST00000586666.1
|
CCDC130
|
coiled-coil domain containing 130 |
| chr2_-_145277569 | 0.07 |
ENST00000303660.4
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr2_-_56150184 | 0.07 |
ENST00000394554.1
|
EFEMP1
|
EGF containing fibulin-like extracellular matrix protein 1 |
| chr11_+_20409227 | 0.07 |
ENST00000437750.2
|
PRMT3
|
protein arginine methyltransferase 3 |
| chr10_+_21823243 | 0.07 |
ENST00000307729.7
ENST00000377091.2 |
MLLT10
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 |
| chr11_+_64002292 | 0.07 |
ENST00000426086.2
|
VEGFB
|
vascular endothelial growth factor B |
| chr17_+_73717551 | 0.07 |
ENST00000450894.3
|
ITGB4
|
integrin, beta 4 |
| chr7_+_131012605 | 0.07 |
ENST00000446815.1
ENST00000352689.6 |
MKLN1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr21_-_36421401 | 0.07 |
ENST00000486278.2
|
RUNX1
|
runt-related transcription factor 1 |
| chr22_+_19939026 | 0.07 |
ENST00000406520.3
|
COMT
|
catechol-O-methyltransferase |
| chr22_+_20067738 | 0.07 |
ENST00000351989.3
ENST00000383024.2 |
DGCR8
|
DGCR8 microprocessor complex subunit |
| chr5_-_180235755 | 0.06 |
ENST00000502678.1
|
MGAT1
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| chr2_+_173940668 | 0.06 |
ENST00000375213.3
|
MLTK
|
Mitogen-activated protein kinase kinase kinase MLT |
| chr7_+_112063192 | 0.06 |
ENST00000005558.4
|
IFRD1
|
interferon-related developmental regulator 1 |
| chr4_+_37455536 | 0.06 |
ENST00000381980.4
ENST00000508175.1 |
C4orf19
|
chromosome 4 open reading frame 19 |
| chr11_-_73720122 | 0.06 |
ENST00000426995.2
|
UCP3
|
uncoupling protein 3 (mitochondrial, proton carrier) |
| chr17_+_73717516 | 0.06 |
ENST00000200181.3
ENST00000339591.3 |
ITGB4
|
integrin, beta 4 |
| chr16_-_84587633 | 0.06 |
ENST00000565079.1
|
TLDC1
|
TBC/LysM-associated domain containing 1 |
| chr18_+_20494078 | 0.06 |
ENST00000579124.1
ENST00000577588.1 ENST00000582354.1 ENST00000581819.1 |
RBBP8
|
retinoblastoma binding protein 8 |
| chr8_-_65730127 | 0.06 |
ENST00000522106.1
|
RP11-1D12.2
|
RP11-1D12.2 |
| chr8_+_356942 | 0.06 |
ENST00000276326.5
|
FBXO25
|
F-box protein 25 |
| chr2_-_220025548 | 0.06 |
ENST00000356853.5
|
NHEJ1
|
nonhomologous end-joining factor 1 |
| chr2_+_226265364 | 0.06 |
ENST00000272907.6
|
NYAP2
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 2 |
| chr6_+_123100853 | 0.06 |
ENST00000356535.4
|
FABP7
|
fatty acid binding protein 7, brain |
| chr13_-_96705624 | 0.06 |
ENST00000376747.3
ENST00000376712.4 ENST00000397618.3 ENST00000376714.3 |
UGGT2
|
UDP-glucose glycoprotein glucosyltransferase 2 |
| chr22_-_30234218 | 0.06 |
ENST00000307790.3
ENST00000542393.1 ENST00000397771.2 |
ASCC2
|
activating signal cointegrator 1 complex subunit 2 |
| chr1_+_165600436 | 0.06 |
ENST00000367888.4
ENST00000367885.1 ENST00000367884.2 |
MGST3
|
microsomal glutathione S-transferase 3 |
| chr10_+_51572339 | 0.06 |
ENST00000344348.6
|
NCOA4
|
nuclear receptor coactivator 4 |
| chr8_-_101118185 | 0.06 |
ENST00000523437.1
|
RGS22
|
regulator of G-protein signaling 22 |
| chr2_+_54683419 | 0.06 |
ENST00000356805.4
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1 |
| chr5_-_174871136 | 0.06 |
ENST00000393752.2
|
DRD1
|
dopamine receptor D1 |
| chr10_+_51572408 | 0.06 |
ENST00000374082.1
|
NCOA4
|
nuclear receptor coactivator 4 |
| chr3_+_124303472 | 0.06 |
ENST00000291478.5
|
KALRN
|
kalirin, RhoGEF kinase |
| chr2_-_56150910 | 0.06 |
ENST00000424836.2
ENST00000438672.1 ENST00000440439.1 ENST00000429909.1 ENST00000424207.1 ENST00000452337.1 ENST00000355426.3 ENST00000439193.1 ENST00000421664.1 |
EFEMP1
|
EGF containing fibulin-like extracellular matrix protein 1 |
| chr10_+_21823079 | 0.06 |
ENST00000377100.3
ENST00000377072.3 ENST00000446906.2 |
MLLT10
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 |
| chr12_+_41221975 | 0.06 |
ENST00000552913.1
|
CNTN1
|
contactin 1 |
| chrX_-_154842538 | 0.06 |
ENST00000369439.4
|
TMLHE
|
trimethyllysine hydroxylase, epsilon |
| chr2_+_196440692 | 0.06 |
ENST00000458054.1
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr1_+_54411715 | 0.05 |
ENST00000371370.3
ENST00000371368.1 |
LRRC42
|
leucine rich repeat containing 42 |
| chr18_-_45457192 | 0.05 |
ENST00000586514.1
ENST00000591214.1 ENST00000589877.1 |
SMAD2
|
SMAD family member 2 |
| chr11_+_89867681 | 0.05 |
ENST00000534061.1
|
NAALAD2
|
N-acetylated alpha-linked acidic dipeptidase 2 |
| chr22_+_24828087 | 0.05 |
ENST00000472248.1
ENST00000436735.1 |
ADORA2A
|
adenosine A2a receptor |
| chr17_+_73717407 | 0.05 |
ENST00000579662.1
|
ITGB4
|
integrin, beta 4 |
| chr5_+_169010638 | 0.05 |
ENST00000265295.4
ENST00000506574.1 ENST00000515224.1 ENST00000508247.1 ENST00000513941.1 |
SPDL1
|
spindle apparatus coiled-coil protein 1 |
| chr22_+_19938419 | 0.05 |
ENST00000412786.1
|
COMT
|
catechol-O-methyltransferase |
| chr18_+_905104 | 0.05 |
ENST00000579794.1
|
ADCYAP1
|
adenylate cyclase activating polypeptide 1 (pituitary) |
| chr1_+_165600083 | 0.05 |
ENST00000367889.3
|
MGST3
|
microsomal glutathione S-transferase 3 |
| chr12_-_96390108 | 0.05 |
ENST00000538703.1
ENST00000261208.3 |
HAL
|
histidine ammonia-lyase |
| chr1_+_156756667 | 0.05 |
ENST00000526188.1
ENST00000454659.1 |
PRCC
|
papillary renal cell carcinoma (translocation-associated) |
| chr4_+_86396321 | 0.05 |
ENST00000503995.1
|
ARHGAP24
|
Rho GTPase activating protein 24 |
| chr6_+_44126545 | 0.05 |
ENST00000532171.1
ENST00000398776.1 ENST00000542245.1 |
CAPN11
|
calpain 11 |
| chr6_+_32937083 | 0.05 |
ENST00000456339.1
|
BRD2
|
bromodomain containing 2 |
| chr17_+_46131843 | 0.05 |
ENST00000577411.1
|
NFE2L1
|
nuclear factor, erythroid 2-like 1 |
| chr16_-_31147020 | 0.05 |
ENST00000568261.1
ENST00000567797.1 ENST00000317508.6 |
PRSS8
|
protease, serine, 8 |
| chr18_-_21852143 | 0.05 |
ENST00000399443.3
|
OSBPL1A
|
oxysterol binding protein-like 1A |
| chr4_+_48833183 | 0.05 |
ENST00000503016.1
|
OCIAD1
|
OCIA domain containing 1 |
| chr11_+_20409070 | 0.05 |
ENST00000331079.6
|
PRMT3
|
protein arginine methyltransferase 3 |
| chr21_-_27542972 | 0.05 |
ENST00000346798.3
ENST00000439274.2 ENST00000354192.3 ENST00000348990.5 ENST00000357903.3 ENST00000358918.3 ENST00000359726.3 |
APP
|
amyloid beta (A4) precursor protein |
| chr7_-_140098257 | 0.05 |
ENST00000340308.3
ENST00000447932.2 ENST00000429996.2 ENST00000469193.1 ENST00000326232.9 |
SLC37A3
|
solute carrier family 37, member 3 |
| chr12_-_14996355 | 0.05 |
ENST00000228936.4
|
ART4
|
ADP-ribosyltransferase 4 (Dombrock blood group) |
| chr10_-_96122682 | 0.05 |
ENST00000371361.3
|
NOC3L
|
nucleolar complex associated 3 homolog (S. cerevisiae) |
| chr1_+_210111534 | 0.05 |
ENST00000422431.1
ENST00000534859.1 ENST00000399639.2 ENST00000537238.1 |
SYT14
|
synaptotagmin XIV |
| chr1_+_20465805 | 0.05 |
ENST00000375102.3
|
PLA2G2F
|
phospholipase A2, group IIF |
| chr2_+_208104497 | 0.05 |
ENST00000430494.1
|
AC007879.7
|
AC007879.7 |
| chr3_+_136581096 | 0.05 |
ENST00000476286.1
ENST00000488930.1 |
NCK1
|
NCK adaptor protein 1 |
| chr17_+_75450099 | 0.05 |
ENST00000586433.1
|
SEPT9
|
septin 9 |
| chr8_+_107460147 | 0.05 |
ENST00000442977.2
|
OXR1
|
oxidation resistance 1 |
| chr2_+_85839218 | 0.05 |
ENST00000448971.1
ENST00000442708.1 ENST00000450066.2 |
USP39
|
ubiquitin specific peptidase 39 |
| chr22_+_21321531 | 0.05 |
ENST00000405089.1
ENST00000335375.5 |
AIFM3
|
apoptosis-inducing factor, mitochondrion-associated, 3 |
| chr3_-_39321512 | 0.04 |
ENST00000399220.2
|
CX3CR1
|
chemokine (C-X3-C motif) receptor 1 |
| chr10_+_8096631 | 0.04 |
ENST00000379328.3
|
GATA3
|
GATA binding protein 3 |
| chr22_-_37640277 | 0.04 |
ENST00000401529.3
ENST00000249071.6 |
RAC2
|
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
| chr11_-_74109422 | 0.04 |
ENST00000298198.4
|
PGM2L1
|
phosphoglucomutase 2-like 1 |
| chr4_+_39046615 | 0.04 |
ENST00000261425.3
ENST00000508137.2 |
KLHL5
|
kelch-like family member 5 |
| chr3_-_122512619 | 0.04 |
ENST00000383659.1
ENST00000306103.2 |
HSPBAP1
|
HSPB (heat shock 27kDa) associated protein 1 |
| chr2_-_201753717 | 0.04 |
ENST00000409264.2
|
PPIL3
|
peptidylprolyl isomerase (cyclophilin)-like 3 |
| chr13_-_52026730 | 0.04 |
ENST00000420668.2
|
INTS6
|
integrator complex subunit 6 |
| chr8_-_101118210 | 0.04 |
ENST00000360863.6
|
RGS22
|
regulator of G-protein signaling 22 |
| chr7_+_128379449 | 0.04 |
ENST00000479257.1
|
CALU
|
calumenin |
| chr1_+_228327923 | 0.04 |
ENST00000391865.3
|
GUK1
|
guanylate kinase 1 |
| chr7_+_30634297 | 0.04 |
ENST00000389266.3
|
GARS
|
glycyl-tRNA synthetase |
| chr17_-_42767092 | 0.04 |
ENST00000588687.1
|
CCDC43
|
coiled-coil domain containing 43 |
| chr5_+_112073544 | 0.04 |
ENST00000257430.4
ENST00000508376.2 |
APC
|
adenomatous polyposis coli |
| chr3_+_33331156 | 0.04 |
ENST00000542085.1
|
FBXL2
|
F-box and leucine-rich repeat protein 2 |
| chr22_+_25202232 | 0.04 |
ENST00000400358.4
ENST00000400359.4 |
SGSM1
|
small G protein signaling modulator 1 |
| chr12_-_48119301 | 0.04 |
ENST00000545824.2
ENST00000422538.3 |
ENDOU
|
endonuclease, polyU-specific |
| chr2_-_145277882 | 0.04 |
ENST00000465070.1
ENST00000444559.1 |
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chr15_+_40531621 | 0.04 |
ENST00000560346.1
|
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6 |
| chr21_+_27543175 | 0.04 |
ENST00000608591.1
ENST00000609365.1 |
AP000230.1
|
AP000230.1 |
| chr20_+_44036900 | 0.04 |
ENST00000443296.1
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr19_-_18709357 | 0.04 |
ENST00000597131.1
|
CRLF1
|
cytokine receptor-like factor 1 |
| chr1_-_25256368 | 0.04 |
ENST00000308873.6
|
RUNX3
|
runt-related transcription factor 3 |
| chr9_-_112260531 | 0.04 |
ENST00000374541.2
ENST00000262539.3 |
PTPN3
|
protein tyrosine phosphatase, non-receptor type 3 |
| chr2_+_85661918 | 0.04 |
ENST00000340326.2
|
SH2D6
|
SH2 domain containing 6 |
| chr20_+_44036620 | 0.04 |
ENST00000372710.3
|
DBNDD2
|
dysbindin (dystrobrevin binding protein 1) domain containing 2 |
| chr4_+_48833119 | 0.04 |
ENST00000444354.2
ENST00000509963.1 ENST00000509246.1 |
OCIAD1
|
OCIA domain containing 1 |
| chr1_-_115292591 | 0.04 |
ENST00000438362.2
|
CSDE1
|
cold shock domain containing E1, RNA-binding |
| chr15_-_55657428 | 0.04 |
ENST00000568543.1
|
CCPG1
|
cell cycle progression 1 |
| chr4_-_6202247 | 0.04 |
ENST00000409021.3
ENST00000409371.3 |
JAKMIP1
|
janus kinase and microtubule interacting protein 1 |
| chr16_-_56458896 | 0.04 |
ENST00000565445.1
|
AMFR
|
autocrine motility factor receptor, E3 ubiquitin protein ligase |
| chr11_+_125439298 | 0.04 |
ENST00000278903.6
ENST00000343678.4 ENST00000524723.1 ENST00000527842.2 |
EI24
|
etoposide induced 2.4 |
| chr5_-_159827073 | 0.04 |
ENST00000408953.3
|
C5orf54
|
chromosome 5 open reading frame 54 |
| chr15_+_40532058 | 0.04 |
ENST00000260404.4
|
PAK6
|
p21 protein (Cdc42/Rac)-activated kinase 6 |
| chr4_-_90758118 | 0.04 |
ENST00000420646.2
|
SNCA
|
synuclein, alpha (non A4 component of amyloid precursor) |
| chr17_+_75450146 | 0.04 |
ENST00000585929.1
ENST00000590917.1 |
SEPT9
|
septin 9 |
| chr2_+_28824786 | 0.04 |
ENST00000541605.1
|
PLB1
|
phospholipase B1 |
| chrX_-_23761317 | 0.04 |
ENST00000492081.1
ENST00000379303.5 ENST00000336430.7 |
ACOT9
|
acyl-CoA thioesterase 9 |
| chr22_+_31003133 | 0.04 |
ENST00000405742.3
|
TCN2
|
transcobalamin II |
| chr11_+_129939811 | 0.04 |
ENST00000345598.5
ENST00000338167.5 |
APLP2
|
amyloid beta (A4) precursor-like protein 2 |
| chr5_+_112074029 | 0.04 |
ENST00000512211.2
|
APC
|
adenomatous polyposis coli |
| chr2_-_134326009 | 0.03 |
ENST00000409261.1
ENST00000409213.1 |
NCKAP5
|
NCK-associated protein 5 |
| chr5_+_75379224 | 0.03 |
ENST00000322285.7
|
SV2C
|
synaptic vesicle glycoprotein 2C |
| chr9_+_116037922 | 0.03 |
ENST00000374198.4
|
PRPF4
|
pre-mRNA processing factor 4 |
| chr1_+_3607228 | 0.03 |
ENST00000378285.1
ENST00000378280.1 ENST00000378288.4 |
TP73
|
tumor protein p73 |
| chr17_+_10600894 | 0.03 |
ENST00000379774.4
|
ADPRM
|
ADP-ribose/CDP-alcohol diphosphatase, manganese-dependent |
| chr8_+_1919481 | 0.03 |
ENST00000518539.2
|
KBTBD11-OT1
|
KBTBD11 overlapping transcript 1 (non-protein coding) |
| chr7_+_45067265 | 0.03 |
ENST00000474617.1
|
CCM2
|
cerebral cavernous malformation 2 |
| chr1_+_205012293 | 0.03 |
ENST00000331830.4
|
CNTN2
|
contactin 2 (axonal) |
| chr11_+_86013253 | 0.03 |
ENST00000533986.1
ENST00000278483.3 |
C11orf73
|
chromosome 11 open reading frame 73 |
| chr8_-_101117847 | 0.03 |
ENST00000523287.1
ENST00000519092.1 |
RGS22
|
regulator of G-protein signaling 22 |
| chrX_-_106243451 | 0.03 |
ENST00000355610.4
ENST00000535534.1 |
MORC4
|
MORC family CW-type zinc finger 4 |
| chr19_+_3880581 | 0.03 |
ENST00000450849.2
ENST00000301260.6 ENST00000398448.3 |
ATCAY
|
ataxia, cerebellar, Cayman type |
| chr13_-_96329048 | 0.03 |
ENST00000606011.1
ENST00000499499.2 |
DNAJC3-AS1
|
DNAJC3 antisense RNA 1 (head to head) |
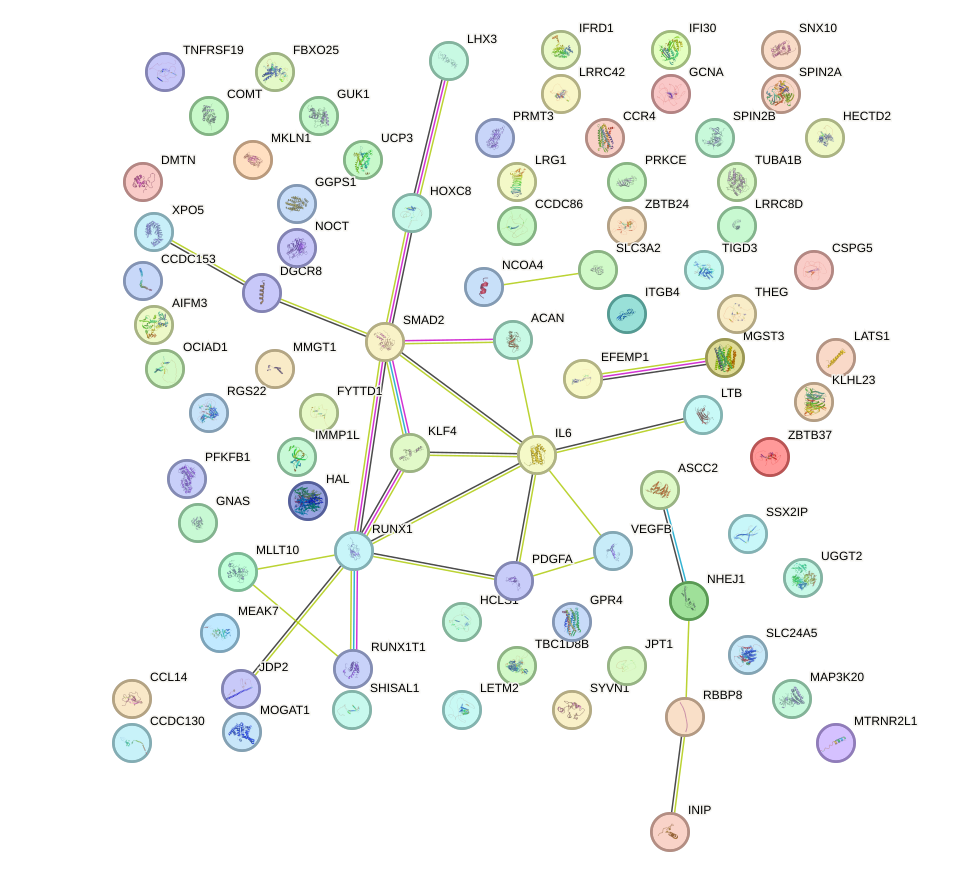
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF784
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:2000470 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.0 | 0.2 | GO:0043606 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.1 | GO:0033385 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.0 | 0.1 | GO:0035281 | pre-miRNA export from nucleus(GO:0035281) |
| 0.0 | 0.4 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0035668 | TRAM-dependent toll-like receptor signaling pathway(GO:0035668) TRAM-dependent toll-like receptor 4 signaling pathway(GO:0035669) |
| 0.0 | 0.3 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 0.1 | GO:0002384 | hepatic immune response(GO:0002384) response to prolactin(GO:1990637) regulation of STAT protein import into nucleus(GO:2000364) positive regulation of STAT protein import into nucleus(GO:2000366) |
| 0.0 | 0.1 | GO:1904995 | negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) |
| 0.0 | 0.1 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.0 | 0.1 | GO:0045963 | negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) |
| 0.0 | 0.2 | GO:0060356 | leucine import(GO:0060356) |
| 0.0 | 0.1 | GO:0009183 | purine deoxyribonucleoside diphosphate biosynthetic process(GO:0009183) |
| 0.0 | 0.1 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.0 | 0.1 | GO:0035054 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.0 | 0.2 | GO:0061373 | mammillary body development(GO:0021767) mammillary axonal complex development(GO:0061373) positive regulation of lens fiber cell differentiation(GO:1902748) regulation of melanosome organization(GO:1903056) |
| 0.0 | 0.1 | GO:0097359 | UDP-glucosylation(GO:0097359) |
| 0.0 | 0.1 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 0.1 | GO:0048050 | post-embryonic eye morphogenesis(GO:0048050) |
| 0.0 | 0.1 | GO:0061289 | cell-cell signaling involved in kidney development(GO:0060995) Wnt signaling pathway involved in kidney development(GO:0061289) canonical Wnt signaling pathway involved in metanephric kidney development(GO:0061290) cell-cell signaling involved in metanephros development(GO:0072204) negative regulation of DNA demethylation(GO:1901536) |
| 0.0 | 0.1 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) inner cell mass cellular morphogenesis(GO:0001828) |
| 0.0 | 0.0 | GO:1903674 | regulation of cap-dependent translational initiation(GO:1903674) positive regulation of cap-dependent translational initiation(GO:1903676) |
| 0.0 | 0.0 | GO:0002881 | negative regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002881) |
| 0.0 | 0.1 | GO:0035188 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.0 | 0.2 | GO:0010579 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.0 | 0.1 | GO:0010512 | negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) |
| 0.0 | 0.3 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.1 | GO:1903301 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.0 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.0 | 0.1 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.0 | 0.1 | GO:1903615 | regulation of protein tyrosine phosphatase activity(GO:1903613) positive regulation of protein tyrosine phosphatase activity(GO:1903615) |
| 0.0 | 0.1 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.2 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.0 | GO:0015966 | diadenosine polyphosphate biosynthetic process(GO:0015960) diadenosine tetraphosphate metabolic process(GO:0015965) diadenosine tetraphosphate biosynthetic process(GO:0015966) |
| 0.0 | 0.1 | GO:0019919 | peptidyl-arginine methylation, to asymmetrical-dimethyl arginine(GO:0019919) |
| 0.0 | 0.0 | GO:0035793 | positive regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:0035793) regulation of metanephric mesenchymal cell migration by platelet-derived growth factor receptor-beta signaling pathway(GO:1900238) positive regulation of metanephric mesenchymal cell migration(GO:2000591) |
| 0.0 | 0.1 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0042565 | RNA nuclear export complex(GO:0042565) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.1 | GO:0070877 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.2 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.1 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.1 | 0.2 | GO:0004397 | histidine ammonia-lyase activity(GO:0004397) |
| 0.0 | 0.1 | GO:0035276 | ethanol binding(GO:0035276) |
| 0.0 | 0.1 | GO:0090631 | pre-miRNA transporter activity(GO:0090631) |
| 0.0 | 0.1 | GO:0004337 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.3 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.1 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.0 | GO:0016495 | C-X3-C chemokine receptor activity(GO:0016495) |
| 0.0 | 0.1 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.1 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.2 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.0 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.1 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 0.1 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.1 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |