Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
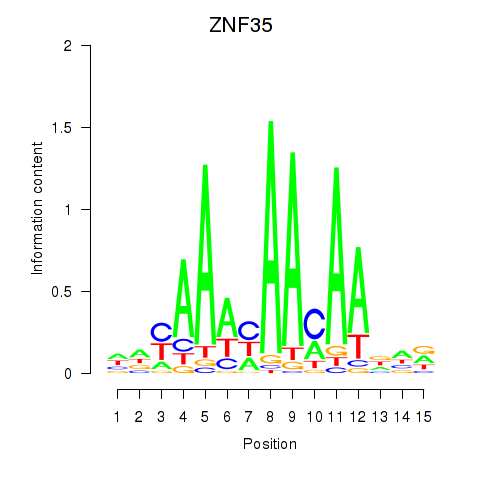
Results for ZNF35
Z-value: 0.13
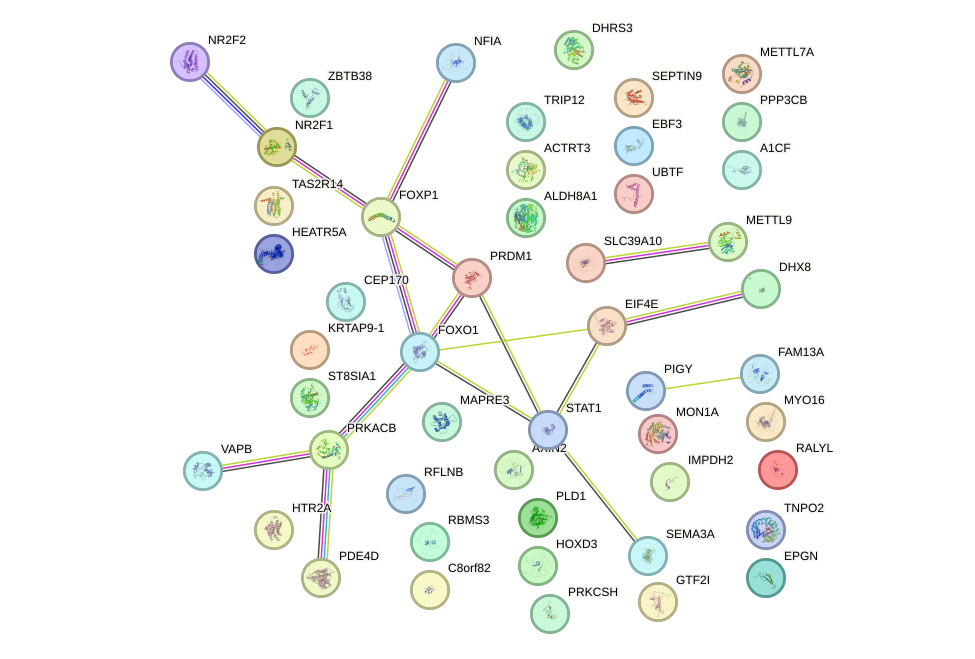
Transcription factors associated with ZNF35
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF35
|
ENSG00000169981.6 | zinc finger protein 35 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF35 | hg19_v2_chr3_+_44690211_44690267 | -0.45 | 5.5e-01 | Click! |
Activity profile of ZNF35 motif
Sorted Z-values of ZNF35 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_145754428 | 0.05 |
ENST00000527462.1
ENST00000313465.5 ENST00000524821.1 |
C8orf82
|
chromosome 8 open reading frame 82 |
| chr2_-_230787879 | 0.04 |
ENST00000435716.1
|
TRIP12
|
thyroid hormone receptor interactor 12 |
| chr3_-_171489085 | 0.04 |
ENST00000418087.1
|
PLD1
|
phospholipase D1, phosphatidylcholine-specific |
| chr9_+_130853715 | 0.03 |
ENST00000373066.5
ENST00000432073.2 |
SLC25A25
|
solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25 |
| chr7_-_83824449 | 0.03 |
ENST00000420047.1
|
SEMA3A
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
| chr12_-_22487618 | 0.03 |
ENST00000404299.3
ENST00000396037.4 |
ST8SIA1
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 |
| chr5_+_72509751 | 0.02 |
ENST00000515556.1
ENST00000513379.1 ENST00000427584.2 |
RP11-60A8.1
|
RP11-60A8.1 |
| chr5_-_39203093 | 0.02 |
ENST00000515010.1
|
FYB
|
FYN binding protein |
| chr5_-_68339648 | 0.02 |
ENST00000479830.2
|
CTC-498J12.1
|
CTC-498J12.1 |
| chr3_-_49967292 | 0.02 |
ENST00000455683.2
|
MON1A
|
MON1 secretory trafficking family member A |
| chr2_+_177015122 | 0.02 |
ENST00000468418.3
|
HOXD3
|
homeobox D3 |
| chr21_+_17792672 | 0.02 |
ENST00000602620.1
|
LINC00478
|
long intergenic non-protein coding RNA 478 |
| chr5_-_58571935 | 0.02 |
ENST00000503258.1
|
PDE4D
|
phosphodiesterase 4D, cAMP-specific |
| chr12_+_51318513 | 0.02 |
ENST00000332160.4
|
METTL7A
|
methyltransferase like 7A |
| chr16_+_21623958 | 0.02 |
ENST00000568826.1
|
METTL9
|
methyltransferase like 9 |
| chr6_+_106535455 | 0.02 |
ENST00000424894.1
|
PRDM1
|
PR domain containing 1, with ZNF domain |
| chr4_+_75174180 | 0.02 |
ENST00000413830.1
|
EPGN
|
epithelial mitogen |
| chr12_-_22487588 | 0.02 |
ENST00000381424.3
|
ST8SIA1
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 |
| chr1_+_84630574 | 0.02 |
ENST00000413538.1
ENST00000417530.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr13_-_47471155 | 0.01 |
ENST00000543956.1
ENST00000542664.1 |
HTR2A
|
5-hydroxytryptamine (serotonin) receptor 2A, G protein-coupled |
| chr4_-_89442940 | 0.01 |
ENST00000527353.1
|
PIGY
|
phosphatidylinositol glycan anchor biosynthesis, class Y |
| chr15_+_96876340 | 0.01 |
ENST00000453270.2
|
NR2F2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr14_-_31889737 | 0.01 |
ENST00000382464.2
|
HEATR5A
|
HEAT repeat containing 5A |
| chr1_+_61869748 | 0.01 |
ENST00000357977.5
|
NFIA
|
nuclear factor I/A |
| chr12_-_11091862 | 0.01 |
ENST00000537503.1
|
TAS2R14
|
taste receptor, type 2, member 14 |
| chr17_-_295730 | 0.01 |
ENST00000329099.4
|
FAM101B
|
family with sequence similarity 101, member B |
| chr3_-_169487617 | 0.01 |
ENST00000330368.2
|
ACTRT3
|
actin-related protein T3 |
| chr13_+_109248500 | 0.01 |
ENST00000356711.2
|
MYO16
|
myosin XVI |
| chr4_-_89744365 | 0.01 |
ENST00000513837.1
ENST00000503556.1 |
FAM13A
|
family with sequence similarity 13, member A |
| chr2_-_191878162 | 0.01 |
ENST00000540176.1
|
STAT1
|
signal transducer and activator of transcription 1, 91kDa |
| chr10_-_4285923 | 0.01 |
ENST00000418372.1
ENST00000608792.1 |
LINC00702
|
long intergenic non-protein coding RNA 702 |
| chr3_-_71632894 | 0.01 |
ENST00000493089.1
|
FOXP1
|
forkhead box P1 |
| chr3_+_141105705 | 0.01 |
ENST00000513258.1
|
ZBTB38
|
zinc finger and BTB domain containing 38 |
| chr8_+_85095769 | 0.01 |
ENST00000518566.1
|
RALYL
|
RALY RNA binding protein-like |
| chr3_-_49066811 | 0.01 |
ENST00000442157.1
ENST00000326739.4 |
IMPDH2
|
IMP (inosine 5'-monophosphate) dehydrogenase 2 |
| chr10_-_52645416 | 0.01 |
ENST00000374001.2
ENST00000373997.3 ENST00000373995.3 ENST00000282641.2 ENST00000395495.1 ENST00000414883.1 |
A1CF
|
APOBEC1 complementation factor |
| chr13_-_41240717 | 0.01 |
ENST00000379561.5
|
FOXO1
|
forkhead box O1 |
| chr7_+_74072288 | 0.01 |
ENST00000443166.1
|
GTF2I
|
general transcription factor IIi |
| chr10_-_131762105 | 0.00 |
ENST00000368648.3
ENST00000355311.5 |
EBF3
|
early B-cell factor 3 |
| chr10_-_75226166 | 0.00 |
ENST00000544628.1
|
PPP3CB
|
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr17_-_63557759 | 0.00 |
ENST00000307078.5
|
AXIN2
|
axin 2 |
| chr18_+_74240610 | 0.00 |
ENST00000578092.1
ENST00000578613.1 ENST00000583578.1 |
LINC00908
|
long intergenic non-protein coding RNA 908 |
| chr17_+_39346139 | 0.00 |
ENST00000398470.1
ENST00000318329.5 |
KRTAP9-1
|
keratin associated protein 9-1 |
| chr20_+_56964169 | 0.00 |
ENST00000475243.1
|
VAPB
|
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr4_-_99850243 | 0.00 |
ENST00000280892.6
ENST00000511644.1 ENST00000504432.1 ENST00000505992.1 |
EIF4E
|
eukaryotic translation initiation factor 4E |
| chr17_+_75369400 | 0.00 |
ENST00000590059.1
|
SEPT9
|
septin 9 |
| chr3_+_69985792 | 0.00 |
ENST00000531774.1
|
MITF
|
microphthalmia-associated transcription factor |
| chr17_-_42295870 | 0.00 |
ENST00000526094.1
ENST00000529383.1 ENST00000530828.1 |
UBTF
|
upstream binding transcription factor, RNA polymerase I |
| chr19_-_12833164 | 0.00 |
ENST00000356861.5
|
TNPO2
|
transportin 2 |
| chr1_-_12677714 | 0.00 |
ENST00000376223.2
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr2_+_196440692 | 0.00 |
ENST00000458054.1
|
SLC39A10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr3_+_29323043 | 0.00 |
ENST00000452462.1
ENST00000456853.1 |
RBMS3
|
RNA binding motif, single stranded interacting protein 3 |
| chr15_+_69857515 | 0.00 |
ENST00000559477.1
|
RP11-279F6.1
|
RP11-279F6.1 |
| chrX_+_9431324 | 0.00 |
ENST00000407597.2
ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X
|
transducin (beta)-like 1X-linked |
| chr3_+_69985734 | 0.00 |
ENST00000314557.6
ENST00000394351.3 |
MITF
|
microphthalmia-associated transcription factor |
| chr13_-_36429763 | 0.00 |
ENST00000379893.1
|
DCLK1
|
doublecortin-like kinase 1 |
| chr6_-_135271219 | 0.00 |
ENST00000367847.2
ENST00000367845.2 |
ALDH8A1
|
aldehyde dehydrogenase 8 family, member A1 |
| chr1_-_243349684 | 0.00 |
ENST00000522895.1
|
CEP170
|
centrosomal protein 170kDa |
| chr21_+_35553045 | 0.00 |
ENST00000416145.1
ENST00000430922.1 ENST00000419881.2 |
LINC00310
|
long intergenic non-protein coding RNA 310 |
| chr17_+_41561317 | 0.00 |
ENST00000540306.1
ENST00000262415.3 ENST00000605777.1 |
DHX8
|
DEAH (Asp-Glu-Ala-His) box polypeptide 8 |