Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
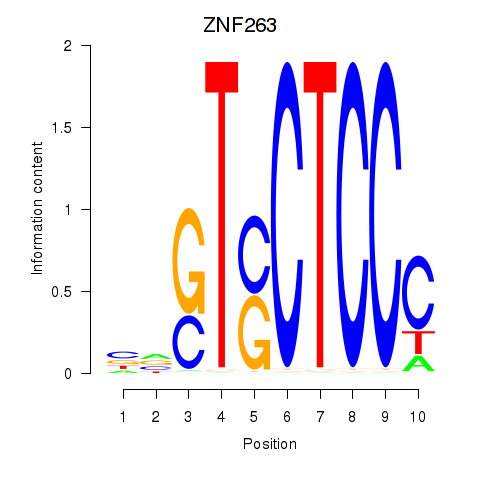
Results for ZNF263
Z-value: 1.17
Transcription factors associated with ZNF263
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZNF263
|
ENSG00000006194.6 | zinc finger protein 263 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZNF263 | hg19_v2_chr16_+_3333443_3333545 | -0.03 | 9.7e-01 | Click! |
Activity profile of ZNF263 motif
Sorted Z-values of ZNF263 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_102268708 | 0.96 |
ENST00000525819.1
|
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr2_-_39664206 | 0.65 |
ENST00000484274.1
|
MAP4K3
|
mitogen-activated protein kinase kinase kinase kinase 3 |
| chr19_-_54618650 | 0.51 |
ENST00000391757.1
|
TFPT
|
TCF3 (E2A) fusion partner (in childhood Leukemia) |
| chr19_-_55658687 | 0.46 |
ENST00000593046.1
|
TNNT1
|
troponin T type 1 (skeletal, slow) |
| chr4_-_84255935 | 0.44 |
ENST00000513463.1
|
HPSE
|
heparanase |
| chr1_+_211432700 | 0.44 |
ENST00000452621.2
|
RCOR3
|
REST corepressor 3 |
| chr17_+_7748233 | 0.39 |
ENST00000570632.1
|
KDM6B
|
lysine (K)-specific demethylase 6B |
| chr10_+_30723533 | 0.39 |
ENST00000413724.1
|
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr16_-_31021717 | 0.39 |
ENST00000565419.1
|
STX1B
|
syntaxin 1B |
| chr17_+_46985823 | 0.37 |
ENST00000508468.2
|
UBE2Z
|
ubiquitin-conjugating enzyme E2Z |
| chr20_-_44455976 | 0.36 |
ENST00000372555.3
|
TNNC2
|
troponin C type 2 (fast) |
| chr21_-_43916433 | 0.36 |
ENST00000291536.3
|
RSPH1
|
radial spoke head 1 homolog (Chlamydomonas) |
| chrX_+_150565038 | 0.36 |
ENST00000370361.1
|
VMA21
|
VMA21 vacuolar H+-ATPase homolog (S. cerevisiae) |
| chr3_+_49044765 | 0.34 |
ENST00000429900.2
|
WDR6
|
WD repeat domain 6 |
| chr15_+_100347228 | 0.34 |
ENST00000559714.1
ENST00000560059.1 |
CTD-2054N24.2
|
Uncharacterized protein |
| chr19_+_55888186 | 0.33 |
ENST00000291934.3
|
TMEM190
|
transmembrane protein 190 |
| chr17_-_27277615 | 0.33 |
ENST00000583747.1
ENST00000584236.1 |
PHF12
|
PHD finger protein 12 |
| chr14_-_27066960 | 0.33 |
ENST00000539517.2
|
NOVA1
|
neuro-oncological ventral antigen 1 |
| chr12_-_80083807 | 0.33 |
ENST00000551712.1
|
PAWR
|
PRKC, apoptosis, WT1, regulator |
| chr11_-_46142615 | 0.33 |
ENST00000529734.1
ENST00000323180.6 |
PHF21A
|
PHD finger protein 21A |
| chr19_+_16178317 | 0.32 |
ENST00000344824.6
ENST00000538887.1 |
TPM4
|
tropomyosin 4 |
| chr8_+_61429728 | 0.32 |
ENST00000529579.1
|
RAB2A
|
RAB2A, member RAS oncogene family |
| chr10_-_46342675 | 0.32 |
ENST00000492347.1
|
AGAP4
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 4 |
| chr7_+_130794878 | 0.31 |
ENST00000416992.2
|
MKLN1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr2_+_149402989 | 0.30 |
ENST00000397424.2
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr12_-_7281469 | 0.30 |
ENST00000542370.1
ENST00000266560.3 |
RBP5
|
retinol binding protein 5, cellular |
| chr9_-_139268068 | 0.30 |
ENST00000371734.3
ENST00000371732.5 ENST00000315908.7 |
CARD9
|
caspase recruitment domain family, member 9 |
| chr12_+_6930964 | 0.30 |
ENST00000382315.3
|
GPR162
|
G protein-coupled receptor 162 |
| chr12_+_54379569 | 0.30 |
ENST00000513209.1
|
RP11-834C11.12
|
RP11-834C11.12 |
| chr17_-_41322332 | 0.30 |
ENST00000590740.1
|
RP11-242D8.1
|
RP11-242D8.1 |
| chr19_+_55587266 | 0.28 |
ENST00000201647.6
ENST00000540810.1 |
EPS8L1
|
EPS8-like 1 |
| chr5_-_81046904 | 0.28 |
ENST00000515395.1
|
SSBP2
|
single-stranded DNA binding protein 2 |
| chr8_-_101322132 | 0.28 |
ENST00000523481.1
|
RNF19A
|
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
| chr15_+_96897466 | 0.27 |
ENST00000558382.1
ENST00000558499.1 |
RP11-522B15.3
|
RP11-522B15.3 |
| chr12_+_6930703 | 0.27 |
ENST00000311268.3
|
GPR162
|
G protein-coupled receptor 162 |
| chr4_-_185395191 | 0.27 |
ENST00000510814.1
ENST00000507523.1 ENST00000506230.1 |
IRF2
|
interferon regulatory factor 2 |
| chr11_+_45168182 | 0.27 |
ENST00000526442.1
|
PRDM11
|
PR domain containing 11 |
| chr7_-_100076765 | 0.27 |
ENST00000393991.1
|
TSC22D4
|
TSC22 domain family, member 4 |
| chr5_+_61602236 | 0.27 |
ENST00000514082.1
ENST00000407818.3 |
KIF2A
|
kinesin heavy chain member 2A |
| chrX_+_69674943 | 0.26 |
ENST00000542398.1
|
DLG3
|
discs, large homolog 3 (Drosophila) |
| chr11_-_130184470 | 0.26 |
ENST00000357899.4
ENST00000397753.1 |
ZBTB44
|
zinc finger and BTB domain containing 44 |
| chr19_+_3224700 | 0.26 |
ENST00000292672.2
ENST00000541430.2 |
CELF5
|
CUGBP, Elav-like family member 5 |
| chr2_-_232328867 | 0.26 |
ENST00000453992.1
ENST00000417652.1 ENST00000454824.1 |
NCL
|
nucleolin |
| chr11_-_129872672 | 0.25 |
ENST00000531431.1
ENST00000527581.1 |
PRDM10
|
PR domain containing 10 |
| chr1_-_85156417 | 0.25 |
ENST00000422026.1
|
SSX2IP
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chr15_-_71146460 | 0.24 |
ENST00000344870.4
|
LARP6
|
La ribonucleoprotein domain family, member 6 |
| chr2_-_74645669 | 0.24 |
ENST00000518401.1
|
C2orf81
|
chromosome 2 open reading frame 81 |
| chr7_+_130794846 | 0.24 |
ENST00000421797.2
|
MKLN1
|
muskelin 1, intracellular mediator containing kelch motifs |
| chr1_+_246729815 | 0.24 |
ENST00000366511.1
|
CNST
|
consortin, connexin sorting protein |
| chr5_+_74807886 | 0.24 |
ENST00000514296.1
|
POLK
|
polymerase (DNA directed) kappa |
| chr4_-_75719896 | 0.24 |
ENST00000395743.3
|
BTC
|
betacellulin |
| chr13_-_52026730 | 0.23 |
ENST00000420668.2
|
INTS6
|
integrator complex subunit 6 |
| chr6_-_111136299 | 0.23 |
ENST00000457688.1
|
CDK19
|
cyclin-dependent kinase 19 |
| chr8_-_125486755 | 0.23 |
ENST00000499418.2
ENST00000530778.1 |
RNF139-AS1
|
RNF139 antisense RNA 1 (head to head) |
| chr20_-_32580924 | 0.23 |
ENST00000432859.1
|
RP5-1125A11.1
|
RP5-1125A11.1 |
| chr5_+_56205878 | 0.23 |
ENST00000423328.1
|
SETD9
|
SET domain containing 9 |
| chr10_+_114710516 | 0.23 |
ENST00000542695.1
ENST00000346198.4 |
TCF7L2
|
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr19_+_48112371 | 0.23 |
ENST00000594866.1
|
GLTSCR1
|
glioma tumor suppressor candidate region gene 1 |
| chr6_-_32157947 | 0.23 |
ENST00000375050.4
|
PBX2
|
pre-B-cell leukemia homeobox 2 |
| chr8_-_92053042 | 0.23 |
ENST00000520014.1
|
TMEM55A
|
transmembrane protein 55A |
| chr13_+_98628886 | 0.22 |
ENST00000490680.1
ENST00000539640.1 ENST00000403772.3 |
IPO5
|
importin 5 |
| chr3_-_171178157 | 0.22 |
ENST00000465393.1
ENST00000436636.2 ENST00000369326.5 ENST00000538048.1 ENST00000341852.6 |
TNIK
|
TRAF2 and NCK interacting kinase |
| chr1_-_225840747 | 0.22 |
ENST00000366843.2
ENST00000366844.3 |
ENAH
|
enabled homolog (Drosophila) |
| chr14_-_51297837 | 0.22 |
ENST00000245441.5
ENST00000389868.3 ENST00000382041.3 ENST00000324330.9 ENST00000453196.1 ENST00000453401.2 |
NIN
|
ninein (GSK3B interacting protein) |
| chr19_-_55658650 | 0.22 |
ENST00000589226.1
|
TNNT1
|
troponin T type 1 (skeletal, slow) |
| chr17_-_58469329 | 0.22 |
ENST00000393003.3
|
USP32
|
ubiquitin specific peptidase 32 |
| chr3_+_195447738 | 0.22 |
ENST00000447234.2
ENST00000320736.6 ENST00000436408.1 |
MUC20
|
mucin 20, cell surface associated |
| chr21_-_43916296 | 0.22 |
ENST00000398352.3
|
RSPH1
|
radial spoke head 1 homolog (Chlamydomonas) |
| chr5_-_74807418 | 0.22 |
ENST00000405807.4
ENST00000261415.7 |
COL4A3BP
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein |
| chr3_+_157823609 | 0.21 |
ENST00000480820.1
|
RSRC1
|
arginine/serine-rich coiled-coil 1 |
| chr3_-_133614297 | 0.21 |
ENST00000486858.1
ENST00000477759.1 |
RAB6B
|
RAB6B, member RAS oncogene family |
| chr3_+_159943362 | 0.21 |
ENST00000326474.3
|
C3orf80
|
chromosome 3 open reading frame 80 |
| chr17_+_65374075 | 0.21 |
ENST00000581322.1
|
PITPNC1
|
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr12_-_96793142 | 0.21 |
ENST00000552262.1
ENST00000551816.1 ENST00000552496.1 |
CDK17
|
cyclin-dependent kinase 17 |
| chr3_+_190231891 | 0.20 |
ENST00000434491.1
ENST00000422940.1 ENST00000317757.3 |
IL1RAP
|
interleukin 1 receptor accessory protein |
| chr14_-_71276211 | 0.20 |
ENST00000381250.4
ENST00000555993.2 |
MAP3K9
|
mitogen-activated protein kinase kinase kinase 9 |
| chrX_-_41782683 | 0.20 |
ENST00000378163.1
ENST00000378154.1 |
CASK
|
calcium/calmodulin-dependent serine protein kinase (MAGUK family) |
| chr18_+_77867177 | 0.20 |
ENST00000560752.1
|
ADNP2
|
ADNP homeobox 2 |
| chr7_+_26438187 | 0.19 |
ENST00000439120.1
ENST00000430548.1 ENST00000421862.1 ENST00000449537.1 ENST00000420774.1 ENST00000418758.2 |
AC004540.5
|
AC004540.5 |
| chr11_-_46142505 | 0.19 |
ENST00000524497.1
ENST00000418153.2 |
PHF21A
|
PHD finger protein 21A |
| chr14_-_21994525 | 0.19 |
ENST00000538754.1
|
SALL2
|
spalt-like transcription factor 2 |
| chr15_+_76030311 | 0.19 |
ENST00000543887.1
|
AC019294.1
|
AC019294.1 |
| chr6_-_18155285 | 0.19 |
ENST00000309983.4
|
TPMT
|
thiopurine S-methyltransferase |
| chr2_-_98612379 | 0.19 |
ENST00000425805.2
|
TMEM131
|
transmembrane protein 131 |
| chr16_-_31085033 | 0.19 |
ENST00000414399.1
|
ZNF668
|
zinc finger protein 668 |
| chr19_-_17559376 | 0.19 |
ENST00000341130.5
|
TMEM221
|
transmembrane protein 221 |
| chr12_+_7055631 | 0.19 |
ENST00000543115.1
ENST00000399448.1 |
PTPN6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr19_-_54663473 | 0.19 |
ENST00000222224.3
|
LENG1
|
leukocyte receptor cluster (LRC) member 1 |
| chr19_-_49568311 | 0.19 |
ENST00000595857.1
ENST00000451356.2 |
NTF4
|
neurotrophin 4 |
| chr1_+_89990378 | 0.19 |
ENST00000449440.1
|
LRRC8B
|
leucine rich repeat containing 8 family, member B |
| chr19_-_55658281 | 0.19 |
ENST00000585321.2
ENST00000587465.2 |
TNNT1
|
troponin T type 1 (skeletal, slow) |
| chrX_-_108868390 | 0.19 |
ENST00000372101.2
|
KCNE1L
|
KCNE1-like |
| chr5_+_118604439 | 0.19 |
ENST00000388882.5
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr2_-_45162783 | 0.19 |
ENST00000432125.2
|
RP11-89K21.1
|
RP11-89K21.1 |
| chr19_-_30199516 | 0.18 |
ENST00000591243.1
|
C19orf12
|
chromosome 19 open reading frame 12 |
| chr14_+_71374433 | 0.18 |
ENST00000439984.3
|
PCNX
|
pecanex homolog (Drosophila) |
| chr3_+_107244229 | 0.18 |
ENST00000456419.1
ENST00000402163.2 |
BBX
|
bobby sox homolog (Drosophila) |
| chr1_+_211432775 | 0.18 |
ENST00000419091.2
|
RCOR3
|
REST corepressor 3 |
| chr8_+_81397846 | 0.18 |
ENST00000379091.4
|
ZBTB10
|
zinc finger and BTB domain containing 10 |
| chr3_+_110790867 | 0.18 |
ENST00000486596.1
ENST00000493615.1 |
PVRL3
|
poliovirus receptor-related 3 |
| chr9_+_140125209 | 0.18 |
ENST00000538474.1
|
SLC34A3
|
solute carrier family 34 (type II sodium/phosphate contransporter), member 3 |
| chr17_+_42015654 | 0.18 |
ENST00000565120.1
|
RP11-527L4.2
|
Uncharacterized protein |
| chr15_-_68497657 | 0.18 |
ENST00000448060.2
ENST00000467889.1 |
CALML4
|
calmodulin-like 4 |
| chr7_-_23053693 | 0.18 |
ENST00000409763.1
ENST00000409923.1 |
FAM126A
|
family with sequence similarity 126, member A |
| chr19_+_44037546 | 0.18 |
ENST00000601282.1
|
ZNF575
|
zinc finger protein 575 |
| chr22_+_29876197 | 0.18 |
ENST00000310624.6
|
NEFH
|
neurofilament, heavy polypeptide |
| chr7_-_150038704 | 0.18 |
ENST00000466675.1
ENST00000482669.1 ENST00000467793.1 ENST00000223271.3 |
RARRES2
|
retinoic acid receptor responder (tazarotene induced) 2 |
| chr1_-_23694794 | 0.18 |
ENST00000374608.3
|
ZNF436
|
zinc finger protein 436 |
| chr1_+_53793885 | 0.18 |
ENST00000445039.2
|
RP4-784A16.5
|
RP4-784A16.5 |
| chr2_+_85646054 | 0.17 |
ENST00000389938.2
|
SH2D6
|
SH2 domain containing 6 |
| chr3_+_149531607 | 0.17 |
ENST00000468648.1
ENST00000459632.1 |
RNF13
|
ring finger protein 13 |
| chr5_+_10564432 | 0.17 |
ENST00000296657.5
|
ANKRD33B
|
ankyrin repeat domain 33B |
| chr8_+_98656693 | 0.17 |
ENST00000519934.1
|
MTDH
|
metadherin |
| chr9_-_34523027 | 0.17 |
ENST00000399775.2
|
ENHO
|
energy homeostasis associated |
| chr2_-_11605966 | 0.17 |
ENST00000307236.4
ENST00000542100.1 ENST00000546212.1 |
E2F6
|
E2F transcription factor 6 |
| chr3_-_37218023 | 0.17 |
ENST00000416425.1
|
LRRFIP2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr3_+_187461442 | 0.17 |
ENST00000450760.1
|
RP11-211G3.2
|
RP11-211G3.2 |
| chr10_+_30723045 | 0.17 |
ENST00000542547.1
ENST00000415139.1 |
MAP3K8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr13_-_52027134 | 0.17 |
ENST00000311234.4
ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6
|
integrator complex subunit 6 |
| chr1_-_226374373 | 0.17 |
ENST00000366812.5
|
ACBD3
|
acyl-CoA binding domain containing 3 |
| chr8_+_90914859 | 0.17 |
ENST00000520659.1
|
OSGIN2
|
oxidative stress induced growth inhibitor family member 2 |
| chr19_-_56048456 | 0.17 |
ENST00000413299.1
|
SBK2
|
SH3 domain binding kinase family, member 2 |
| chr17_-_62503015 | 0.17 |
ENST00000581806.1
|
DDX5
|
DEAD (Asp-Glu-Ala-Asp) box helicase 5 |
| chr14_+_23776167 | 0.17 |
ENST00000554635.1
ENST00000557008.1 |
BCL2L2
BCL2L2-PABPN1
|
BCL2-like 2 BCL2L2-PABPN1 readthrough |
| chr19_-_51071302 | 0.17 |
ENST00000389201.3
ENST00000600381.1 |
LRRC4B
|
leucine rich repeat containing 4B |
| chr17_+_41177220 | 0.17 |
ENST00000587250.2
ENST00000544533.1 |
RND2
|
Rho family GTPase 2 |
| chr15_-_32162833 | 0.16 |
ENST00000560598.1
|
OTUD7A
|
OTU domain containing 7A |
| chrX_+_150151824 | 0.16 |
ENST00000455596.1
ENST00000448905.2 |
HMGB3
|
high mobility group box 3 |
| chr1_+_154193643 | 0.16 |
ENST00000456325.1
|
UBAP2L
|
ubiquitin associated protein 2-like |
| chr12_-_25403737 | 0.16 |
ENST00000256078.4
ENST00000556131.1 ENST00000311936.3 ENST00000557334.1 |
KRAS
|
Kirsten rat sarcoma viral oncogene homolog |
| chr17_-_42296855 | 0.16 |
ENST00000436088.1
|
UBTF
|
upstream binding transcription factor, RNA polymerase I |
| chr17_-_56591321 | 0.16 |
ENST00000583243.1
|
MTMR4
|
myotubularin related protein 4 |
| chr17_+_7533439 | 0.16 |
ENST00000441599.2
ENST00000380450.4 ENST00000416273.3 ENST00000575903.1 ENST00000576830.1 ENST00000571153.1 ENST00000575618.1 ENST00000576152.1 |
SHBG
|
sex hormone-binding globulin |
| chr15_+_83776324 | 0.16 |
ENST00000379390.6
ENST00000379386.4 ENST00000565774.1 ENST00000565982.1 |
TM6SF1
|
transmembrane 6 superfamily member 1 |
| chr5_-_41510725 | 0.16 |
ENST00000328457.3
|
PLCXD3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr6_-_31088214 | 0.16 |
ENST00000376288.2
|
CDSN
|
corneodesmosin |
| chr1_+_211432593 | 0.16 |
ENST00000367006.4
|
RCOR3
|
REST corepressor 3 |
| chr17_-_42297092 | 0.16 |
ENST00000393606.3
|
UBTF
|
upstream binding transcription factor, RNA polymerase I |
| chr9_+_130911770 | 0.16 |
ENST00000372998.1
|
LCN2
|
lipocalin 2 |
| chr12_-_91573132 | 0.16 |
ENST00000550563.1
ENST00000546370.1 |
DCN
|
decorin |
| chrX_-_19905577 | 0.16 |
ENST00000379697.3
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr2_+_10091783 | 0.16 |
ENST00000324883.5
|
GRHL1
|
grainyhead-like 1 (Drosophila) |
| chr5_+_56205081 | 0.16 |
ENST00000285947.2
ENST00000541720.1 |
SETD9
|
SET domain containing 9 |
| chr17_+_30264014 | 0.16 |
ENST00000322652.5
ENST00000580398.1 |
SUZ12
|
SUZ12 polycomb repressive complex 2 subunit |
| chr1_+_178482262 | 0.16 |
ENST00000367641.3
ENST00000367639.1 |
TEX35
|
testis expressed 35 |
| chrX_-_67653291 | 0.15 |
ENST00000540071.1
|
OPHN1
|
oligophrenin 1 |
| chr12_-_57030096 | 0.15 |
ENST00000549506.1
|
BAZ2A
|
bromodomain adjacent to zinc finger domain, 2A |
| chr4_-_151936416 | 0.15 |
ENST00000510413.1
ENST00000507224.1 |
LRBA
|
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr14_+_21569245 | 0.15 |
ENST00000556585.2
|
TMEM253
|
transmembrane protein 253 |
| chr9_-_35689900 | 0.15 |
ENST00000378300.5
ENST00000329305.2 ENST00000360958.2 |
TPM2
|
tropomyosin 2 (beta) |
| chrX_-_55187531 | 0.15 |
ENST00000489298.1
ENST00000477847.2 |
FAM104B
|
family with sequence similarity 104, member B |
| chr11_-_96123022 | 0.15 |
ENST00000542662.1
|
CCDC82
|
coiled-coil domain containing 82 |
| chr13_+_76210448 | 0.15 |
ENST00000377499.5
|
LMO7
|
LIM domain 7 |
| chr19_-_54984354 | 0.15 |
ENST00000301200.2
|
CDC42EP5
|
CDC42 effector protein (Rho GTPase binding) 5 |
| chr14_+_77564701 | 0.15 |
ENST00000557115.1
|
KIAA1737
|
CLOCK-interacting pacemaker |
| chr19_-_12912657 | 0.15 |
ENST00000301522.2
|
PRDX2
|
peroxiredoxin 2 |
| chr19_-_2721336 | 0.15 |
ENST00000588128.1
|
DIRAS1
|
DIRAS family, GTP-binding RAS-like 1 |
| chr4_+_141677577 | 0.15 |
ENST00000609937.1
|
RP11-102N12.3
|
RP11-102N12.3 |
| chr6_-_150039170 | 0.15 |
ENST00000458696.2
ENST00000392273.3 |
LATS1
|
large tumor suppressor kinase 1 |
| chr1_-_33168336 | 0.15 |
ENST00000373484.3
|
SYNC
|
syncoilin, intermediate filament protein |
| chr6_-_80657292 | 0.14 |
ENST00000369816.4
|
ELOVL4
|
ELOVL fatty acid elongase 4 |
| chr3_-_50605150 | 0.14 |
ENST00000357203.3
|
C3orf18
|
chromosome 3 open reading frame 18 |
| chr5_-_81046841 | 0.14 |
ENST00000509013.2
ENST00000505980.1 ENST00000509053.1 |
SSBP2
|
single-stranded DNA binding protein 2 |
| chr10_-_25305011 | 0.14 |
ENST00000331161.4
ENST00000376363.1 |
ENKUR
|
enkurin, TRPC channel interacting protein |
| chr5_+_74807581 | 0.14 |
ENST00000241436.4
ENST00000352007.5 |
POLK
|
polymerase (DNA directed) kappa |
| chr16_-_54963026 | 0.14 |
ENST00000560208.1
ENST00000557792.1 |
CRNDE
|
colorectal neoplasia differentially expressed (non-protein coding) |
| chr14_-_27066636 | 0.14 |
ENST00000267422.7
ENST00000344429.5 ENST00000574031.1 ENST00000465357.2 ENST00000547619.1 |
NOVA1
|
neuro-oncological ventral antigen 1 |
| chr13_-_48575376 | 0.14 |
ENST00000434484.1
|
SUCLA2
|
succinate-CoA ligase, ADP-forming, beta subunit |
| chr14_-_102605983 | 0.14 |
ENST00000334701.7
|
HSP90AA1
|
heat shock protein 90kDa alpha (cytosolic), class A member 1 |
| chr17_+_2207238 | 0.14 |
ENST00000575840.1
ENST00000576620.1 ENST00000576848.1 ENST00000574987.1 |
SRR
|
serine racemase |
| chr19_-_663147 | 0.14 |
ENST00000606702.1
|
RNF126
|
ring finger protein 126 |
| chr1_+_205473865 | 0.14 |
ENST00000506215.1
ENST00000419301.1 |
CDK18
|
cyclin-dependent kinase 18 |
| chr3_-_182703688 | 0.14 |
ENST00000466812.1
ENST00000487822.1 ENST00000460412.1 ENST00000469954.1 |
DCUN1D1
|
DCN1, defective in cullin neddylation 1, domain containing 1 |
| chr5_-_41510656 | 0.14 |
ENST00000377801.3
|
PLCXD3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr2_+_42721689 | 0.14 |
ENST00000405592.1
|
MTA3
|
metastasis associated 1 family, member 3 |
| chr13_-_46626847 | 0.14 |
ENST00000242848.4
ENST00000282007.3 |
ZC3H13
|
zinc finger CCCH-type containing 13 |
| chr1_-_41950342 | 0.14 |
ENST00000372587.4
|
EDN2
|
endothelin 2 |
| chr1_+_60280458 | 0.14 |
ENST00000455990.1
ENST00000371208.3 |
HOOK1
|
hook microtubule-tethering protein 1 |
| chr17_-_58469687 | 0.14 |
ENST00000590133.1
|
USP32
|
ubiquitin specific peptidase 32 |
| chr19_+_44488330 | 0.14 |
ENST00000591532.1
ENST00000407951.2 ENST00000270014.2 ENST00000590615.1 ENST00000586454.1 |
ZNF155
|
zinc finger protein 155 |
| chr13_+_111805980 | 0.14 |
ENST00000491775.1
ENST00000466143.1 ENST00000544132.1 |
ARHGEF7
|
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr8_-_54934708 | 0.14 |
ENST00000520534.1
ENST00000518784.1 ENST00000522635.1 |
TCEA1
|
transcription elongation factor A (SII), 1 |
| chr1_+_179923873 | 0.14 |
ENST00000367607.3
ENST00000491495.2 |
CEP350
|
centrosomal protein 350kDa |
| chr1_+_150898733 | 0.14 |
ENST00000525956.1
|
SETDB1
|
SET domain, bifurcated 1 |
| chr13_-_108518986 | 0.14 |
ENST00000375915.2
|
FAM155A
|
family with sequence similarity 155, member A |
| chr3_-_50605077 | 0.14 |
ENST00000426034.1
ENST00000441239.1 |
C3orf18
|
chromosome 3 open reading frame 18 |
| chr7_-_139876734 | 0.14 |
ENST00000006967.5
|
JHDM1D
|
lysine (K)-specific demethylase 7A |
| chr11_+_71259466 | 0.13 |
ENST00000528743.2
|
KRTAP5-9
|
keratin associated protein 5-9 |
| chr3_+_133292851 | 0.13 |
ENST00000503932.1
|
CDV3
|
CDV3 homolog (mouse) |
| chr15_+_49462397 | 0.13 |
ENST00000396509.2
|
GALK2
|
galactokinase 2 |
| chr17_+_37821593 | 0.13 |
ENST00000578283.1
|
TCAP
|
titin-cap |
| chr3_+_133292759 | 0.13 |
ENST00000431519.2
|
CDV3
|
CDV3 homolog (mouse) |
| chrX_-_41782592 | 0.13 |
ENST00000378158.1
|
CASK
|
calcium/calmodulin-dependent serine protein kinase (MAGUK family) |
| chr11_+_111808119 | 0.13 |
ENST00000531396.1
|
DIXDC1
|
DIX domain containing 1 |
| chr6_+_76311636 | 0.13 |
ENST00000327284.8
|
SENP6
|
SUMO1/sentrin specific peptidase 6 |
| chr19_+_57901208 | 0.13 |
ENST00000366197.5
ENST00000596282.1 ENST00000597400.1 ENST00000598895.1 ENST00000336128.7 ENST00000596617.1 |
ZNF548
AC003002.6
|
zinc finger protein 548 Uncharacterized protein |
| chr4_+_38869298 | 0.13 |
ENST00000510213.1
ENST00000515037.1 |
FAM114A1
|
family with sequence similarity 114, member A1 |
| chr12_-_48099754 | 0.13 |
ENST00000380650.4
|
RPAP3
|
RNA polymerase II associated protein 3 |
| chr11_-_73309112 | 0.13 |
ENST00000450446.2
|
FAM168A
|
family with sequence similarity 168, member A |
| chr17_-_9725388 | 0.13 |
ENST00000399363.4
|
RP11-477N12.3
|
Putative germ cell-specific gene 1-like protein 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZNF263
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0010615 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) positive regulation of connective tissue replacement(GO:1905205) |
| 0.1 | 0.9 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.1 | 0.6 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.1 | 0.4 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 0.1 | 0.2 | GO:0090222 | centrosome-templated microtubule nucleation(GO:0090222) |
| 0.1 | 0.2 | GO:0035621 | ER to Golgi ceramide transport(GO:0035621) |
| 0.1 | 0.5 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.1 | 0.3 | GO:0021897 | forebrain astrocyte differentiation(GO:0021896) forebrain astrocyte development(GO:0021897) |
| 0.1 | 0.3 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.1 | 0.2 | GO:0035261 | external genitalia morphogenesis(GO:0035261) |
| 0.1 | 0.2 | GO:0048936 | neurofilament bundle assembly(GO:0033693) peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.1 | 0.2 | GO:0061760 | antifungal humoral response(GO:0019732) antifungal innate immune response(GO:0061760) |
| 0.1 | 0.2 | GO:0008052 | sensory organ boundary specification(GO:0008052) formation of organ boundary(GO:0010160) taste bud development(GO:0061193) |
| 0.1 | 0.2 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.1 | 0.2 | GO:1905053 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.1 | 0.3 | GO:0042998 | positive regulation of Golgi to plasma membrane protein transport(GO:0042998) |
| 0.1 | 0.3 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.1 | 0.2 | GO:0006781 | succinyl-CoA pathway(GO:0006781) |
| 0.1 | 0.3 | GO:0031585 | regulation of inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0031585) |
| 0.1 | 0.3 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.0 | 0.4 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.1 | GO:0043406 | positive regulation of MAP kinase activity(GO:0043406) |
| 0.0 | 0.1 | GO:0071629 | cytoplasm-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071629) |
| 0.0 | 0.1 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.1 | GO:0030241 | skeletal muscle myosin thick filament assembly(GO:0030241) |
| 0.0 | 0.4 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.0 | 0.5 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.1 | GO:0038162 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) erythropoietin-mediated signaling pathway(GO:0038162) |
| 0.0 | 0.0 | GO:0060966 | regulation of posttranscriptional gene silencing(GO:0060147) regulation of gene silencing by miRNA(GO:0060964) regulation of gene silencing by RNA(GO:0060966) |
| 0.0 | 0.2 | GO:0003335 | corneocyte development(GO:0003335) |
| 0.0 | 0.1 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.0 | 0.2 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.0 | 0.3 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 0.2 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.1 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.0 | 0.2 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 0.2 | GO:0001828 | inner cell mass cell fate commitment(GO:0001827) inner cell mass cellular morphogenesis(GO:0001828) |
| 0.0 | 0.0 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.0 | 0.2 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.3 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.0 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.3 | GO:0070235 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.3 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.0 | 0.1 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.0 | 0.3 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.2 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.1 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) |
| 0.0 | 0.1 | GO:0006679 | glucosylceramide biosynthetic process(GO:0006679) |
| 0.0 | 0.1 | GO:0035674 | tricarboxylic acid transmembrane transport(GO:0035674) |
| 0.0 | 0.2 | GO:1902730 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) myoblast fate commitment(GO:0048625) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.1 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.2 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.0 | 0.1 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.1 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.0 | 0.2 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.1 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.0 | 0.1 | GO:0021589 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.0 | 0.1 | GO:1904404 | cellular response to vitamin B1(GO:0071301) response to formaldehyde(GO:1904404) |
| 0.0 | 0.4 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.2 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.3 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.1 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.3 | GO:1902659 | regulation of glucose mediated signaling pathway(GO:1902659) |
| 0.0 | 0.2 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.0 | 0.1 | GO:0014824 | artery smooth muscle contraction(GO:0014824) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.4 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.2 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.2 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.0 | 0.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0072369 | regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.0 | 0.3 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.3 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 0.1 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:0048611 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) right lung development(GO:0060458) |
| 0.0 | 0.1 | GO:0032079 | positive regulation of endodeoxyribonuclease activity(GO:0032079) |
| 0.0 | 0.1 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.3 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.0 | GO:0034085 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.1 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.0 | 0.0 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) |
| 0.0 | 0.2 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.2 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.1 | GO:0033504 | floor plate development(GO:0033504) |
| 0.0 | 0.2 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.1 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) |
| 0.0 | 0.1 | GO:0008291 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.0 | 0.4 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.1 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.0 | 0.1 | GO:0009447 | putrescine catabolic process(GO:0009447) |
| 0.0 | 0.1 | GO:0055096 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 0.0 | 0.1 | GO:1904237 | regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904235) positive regulation of substrate-dependent cell migration, cell attachment to substrate(GO:1904237) |
| 0.0 | 0.1 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.1 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.0 | 0.1 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.1 | GO:0097069 | cellular response to thyroxine stimulus(GO:0097069) cellular response to L-phenylalanine derivative(GO:1904387) |
| 0.0 | 0.0 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.0 | 0.1 | GO:0061075 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) cerebral cortex GABAergic interneuron fate commitment(GO:0021893) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) regulation of amacrine cell differentiation(GO:1902869) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.0 | 0.9 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 0.1 | GO:0006173 | dADP biosynthetic process(GO:0006173) |
| 0.0 | 0.5 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:1902283 | negative regulation of primary amine oxidase activity(GO:1902283) |
| 0.0 | 0.1 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.1 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.1 | GO:0061053 | somite development(GO:0061053) |
| 0.0 | 0.0 | GO:0002881 | negative regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002881) |
| 0.0 | 0.3 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.0 | 0.0 | GO:1903280 | negative regulation of calcium:sodium antiporter activity(GO:1903280) |
| 0.0 | 0.1 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.0 | 0.0 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.1 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.3 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.1 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.0 | 0.2 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 | 0.2 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.0 | 0.0 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.0 | 0.3 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.4 | GO:0035810 | positive regulation of urine volume(GO:0035810) |
| 0.0 | 0.1 | GO:0003131 | mesodermal-endodermal cell signaling(GO:0003131) programmed DNA elimination(GO:0031049) chromosome breakage(GO:0031052) histone H2A-S139 phosphorylation(GO:0035978) positive regulation of cellular response to X-ray(GO:2000685) |
| 0.0 | 0.1 | GO:1904338 | regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.0 | 0.1 | GO:0035860 | glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.0 | 0.2 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.0 | 0.1 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.2 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.2 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.2 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.2 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:0072752 | cellular response to rapamycin(GO:0072752) |
| 0.0 | 0.1 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.0 | 0.1 | GO:0034351 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.0 | 0.3 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 | 0.0 | GO:0032755 | positive regulation of interleukin-6 production(GO:0032755) |
| 0.0 | 0.2 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.0 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.0 | 0.4 | GO:0016180 | snRNA processing(GO:0016180) |
| 0.0 | 0.0 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.0 | 0.1 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.0 | 0.1 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.0 | 0.1 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.1 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.0 | 0.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.1 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.0 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.0 | 0.2 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.0 | 0.1 | GO:0010994 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.0 | 0.2 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.0 | GO:0043132 | NAD transport(GO:0043132) |
| 0.0 | 0.0 | GO:0032097 | positive regulation of response to food(GO:0032097) positive regulation of appetite(GO:0032100) |
| 0.0 | 0.1 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.2 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.0 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 0.0 | 0.0 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.0 | GO:1905246 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.0 | 0.0 | GO:0002276 | basophil activation involved in immune response(GO:0002276) basophil activation(GO:0045575) |
| 0.0 | 0.1 | GO:0051549 | positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 0.1 | GO:0042733 | embryonic digit morphogenesis(GO:0042733) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.0 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) |
| 0.0 | 0.1 | GO:0007501 | mesodermal cell fate specification(GO:0007501) |
| 0.0 | 0.1 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) positive regulation of skeletal muscle cell proliferation(GO:0014858) positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 0.0 | 0.1 | GO:0070673 | response to interleukin-18(GO:0070673) |
| 0.0 | 0.1 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 | 0.0 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.1 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.0 | 0.1 | GO:0048298 | regulation of isotype switching to IgA isotypes(GO:0048296) positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 | 0.2 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.2 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.1 | GO:0010710 | regulation of collagen catabolic process(GO:0010710) |
| 0.0 | 0.1 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.0 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.0 | 0.0 | GO:0097107 | postsynaptic density organization(GO:0097106) postsynaptic density assembly(GO:0097107) gephyrin clustering involved in postsynaptic density assembly(GO:0097116) excitatory synapse assembly(GO:1904861) |
| 0.0 | 0.1 | GO:1902373 | negative regulation of mRNA catabolic process(GO:1902373) |
| 0.0 | 0.1 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.0 | GO:0030573 | bile acid catabolic process(GO:0030573) |
| 0.0 | 0.1 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.1 | GO:2000637 | positive regulation of gene silencing by miRNA(GO:2000637) |
| 0.0 | 0.1 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.1 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.0 | 0.2 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.6 | GO:0035082 | axoneme assembly(GO:0035082) |
| 0.0 | 0.0 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.3 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.1 | 0.9 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 0.4 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 0.6 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.3 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.4 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.1 | GO:0070685 | macropinocytic cup(GO:0070685) |
| 0.0 | 0.4 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.2 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.1 | GO:1990075 | kinesin II complex(GO:0016939) periciliary membrane compartment(GO:1990075) |
| 0.0 | 0.1 | GO:1990015 | mesaxon(GO:0097453) ensheathing process(GO:1990015) |
| 0.0 | 0.2 | GO:1990907 | beta-catenin-TCF7L2 complex(GO:0070369) beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.4 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.1 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.1 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.0 | 0.5 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.2 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.3 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.3 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.1 | GO:0034677 | integrin alpha7-beta1 complex(GO:0034677) |
| 0.0 | 0.1 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.2 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.0 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.0 | 0.1 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.4 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.3 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.4 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.6 | GO:0070822 | Sin3-type complex(GO:0070822) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.5 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.2 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.2 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.1 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.0 | 0.0 | GO:0036457 | keratohyalin granule(GO:0036457) |
| 0.0 | 0.6 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.2 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.0 | GO:0044218 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 0.1 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.1 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.2 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.1 | GO:0071547 | piP-body(GO:0071547) |
| 0.0 | 0.3 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.2 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.0 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 0.0 | 0.1 | GO:0034753 | nuclear aryl hydrocarbon receptor complex(GO:0034753) |
| 0.0 | 0.1 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.1 | 0.9 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.1 | 0.2 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.1 | 0.2 | GO:0031859 | platelet activating factor receptor binding(GO:0031859) |
| 0.1 | 0.2 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.1 | 0.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.4 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.1 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) racemase and epimerase activity, acting on amino acids and derivatives(GO:0016855) racemase activity, acting on amino acids and derivatives(GO:0036361) amino-acid racemase activity(GO:0047661) |
| 0.0 | 0.1 | GO:0031705 | bombesin receptor binding(GO:0031705) |
| 0.0 | 0.4 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 1.0 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.2 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.2 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 0.6 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.4 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.0 | 0.3 | GO:0001165 | RNA polymerase I upstream control element sequence-specific DNA binding(GO:0001165) |
| 0.0 | 0.1 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.0 | 0.3 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.0 | 0.1 | GO:1904928 | coreceptor activity involved in canonical Wnt signaling pathway(GO:1904928) |
| 0.0 | 0.3 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.1 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.0 | 0.3 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.2 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.1 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.0 | 0.2 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.2 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.1 | GO:0017153 | sodium:dicarboxylate symporter activity(GO:0017153) |
| 0.0 | 0.1 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |
| 0.0 | 0.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.3 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.2 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.1 | GO:0016435 | rRNA (guanine) methyltransferase activity(GO:0016435) |
| 0.0 | 0.1 | GO:0047280 | nicotinamide phosphoribosyltransferase activity(GO:0047280) |
| 0.0 | 0.2 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.2 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.0 | GO:0047787 | delta4-3-oxosteroid 5beta-reductase activity(GO:0047787) |
| 0.0 | 0.8 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.3 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.3 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.2 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.1 | GO:0004119 | cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) |
| 0.0 | 0.1 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.1 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.0 | 0.2 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.5 | GO:0001222 | transcription corepressor binding(GO:0001222) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.0 | 0.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.3 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 0.1 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 0.7 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 0.3 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.1 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 0.0 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.1 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0035034 | histone acetyltransferase regulator activity(GO:0035034) |
| 0.0 | 0.1 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.0 | 0.0 | GO:0016495 | C-X3-C chemokine receptor activity(GO:0016495) |
| 0.0 | 0.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.0 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) |
| 0.0 | 0.3 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.2 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.1 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.3 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.6 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.3 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.1 | GO:0070717 | poly-purine tract binding(GO:0070717) |
| 0.0 | 0.1 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.0 | GO:0015633 | zinc transporting ATPase activity(GO:0015633) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.1 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.3 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.0 | 0.2 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.1 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.0 | 0.1 | GO:0052593 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.0 | 0.1 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.1 | GO:0033592 | RNA strand annealing activity(GO:0033592) |
| 0.0 | 0.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.0 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.0 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.2 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.0 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.0 | GO:0097158 | pre-mRNA intronic pyrimidine-rich binding(GO:0097158) |
| 0.0 | 0.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.2 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.0 | GO:0004145 | diamine N-acetyltransferase activity(GO:0004145) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.1 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 0.1 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.6 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 0.0 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 1.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.3 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.5 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.3 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.1 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.7 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.4 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.2 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.2 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |