Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
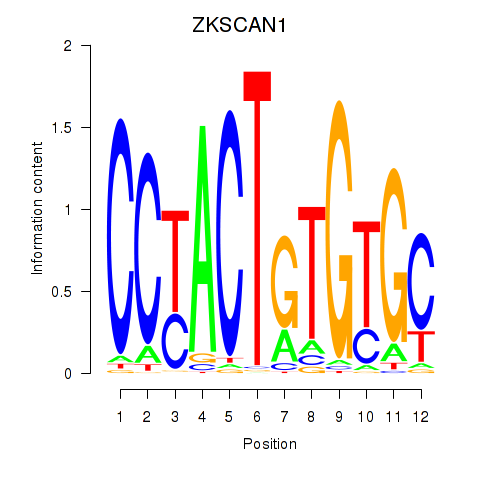
Results for ZKSCAN1
Z-value: 0.44
Transcription factors associated with ZKSCAN1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZKSCAN1
|
ENSG00000106261.12 | zinc finger with KRAB and SCAN domains 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZKSCAN1 | hg19_v2_chr7_+_99613195_99613206 | 0.67 | 3.3e-01 | Click! |
Activity profile of ZKSCAN1 motif
Sorted Z-values of ZKSCAN1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_4364762 | 0.31 |
ENST00000262366.3
|
GLIS2
|
GLIS family zinc finger 2 |
| chr5_+_135394840 | 0.27 |
ENST00000503087.1
|
TGFBI
|
transforming growth factor, beta-induced, 68kDa |
| chr22_+_29702572 | 0.24 |
ENST00000407647.2
ENST00000416823.1 ENST00000428622.1 |
GAS2L1
|
growth arrest-specific 2 like 1 |
| chr14_+_101361107 | 0.21 |
ENST00000553584.1
ENST00000554852.1 |
MEG8
|
maternally expressed 8 (non-protein coding) |
| chr5_+_174151536 | 0.20 |
ENST00000239243.6
ENST00000507785.1 |
MSX2
|
msh homeobox 2 |
| chr6_-_43276535 | 0.19 |
ENST00000372569.3
ENST00000274990.4 |
CRIP3
|
cysteine-rich protein 3 |
| chr16_-_838329 | 0.18 |
ENST00000563560.1
ENST00000569601.1 ENST00000565809.1 ENST00000565377.1 ENST00000007264.2 ENST00000567114.1 |
RPUSD1
|
RNA pseudouridylate synthase domain containing 1 |
| chr17_+_36858694 | 0.17 |
ENST00000563897.1
|
CTB-58E17.1
|
CTB-58E17.1 |
| chr9_+_132934835 | 0.16 |
ENST00000372398.3
|
NCS1
|
neuronal calcium sensor 1 |
| chr19_-_3062465 | 0.15 |
ENST00000327141.4
|
AES
|
amino-terminal enhancer of split |
| chr22_+_30805086 | 0.14 |
ENST00000439838.1
ENST00000439023.3 |
RP4-539M6.19
|
Uncharacterized protein |
| chr1_+_160321120 | 0.12 |
ENST00000424754.1
|
NCSTN
|
nicastrin |
| chr1_+_16083098 | 0.11 |
ENST00000496928.2
ENST00000508310.1 |
FBLIM1
|
filamin binding LIM protein 1 |
| chr1_+_16083123 | 0.11 |
ENST00000510393.1
ENST00000430076.1 |
FBLIM1
|
filamin binding LIM protein 1 |
| chr20_-_56265680 | 0.11 |
ENST00000414037.1
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1 |
| chr16_+_616995 | 0.11 |
ENST00000293874.2
ENST00000409527.2 ENST00000424439.2 ENST00000540585.1 |
PIGQ
NHLRC4
|
phosphatidylinositol glycan anchor biosynthesis, class Q NHL repeat containing 4 |
| chrX_+_47441712 | 0.11 |
ENST00000218388.4
ENST00000377018.2 ENST00000456754.2 ENST00000377017.1 ENST00000441738.1 |
TIMP1
|
TIMP metallopeptidase inhibitor 1 |
| chr1_+_16083154 | 0.11 |
ENST00000375771.1
|
FBLIM1
|
filamin binding LIM protein 1 |
| chr17_+_18163848 | 0.11 |
ENST00000323019.4
ENST00000578174.1 ENST00000395704.4 ENST00000395703.4 ENST00000578621.1 ENST00000579341.1 |
MIEF2
|
mitochondrial elongation factor 2 |
| chr19_-_3063099 | 0.10 |
ENST00000221561.8
|
AES
|
amino-terminal enhancer of split |
| chr1_+_27648648 | 0.10 |
ENST00000374076.4
|
TMEM222
|
transmembrane protein 222 |
| chr20_+_4152356 | 0.10 |
ENST00000379460.2
|
SMOX
|
spermine oxidase |
| chr19_+_8478154 | 0.10 |
ENST00000381035.4
ENST00000595142.1 ENST00000601724.1 ENST00000393944.1 ENST00000215555.2 ENST00000601283.1 ENST00000595213.1 |
MARCH2
|
membrane-associated ring finger (C3HC4) 2, E3 ubiquitin protein ligase |
| chr19_-_2739992 | 0.09 |
ENST00000545664.1
ENST00000589363.1 ENST00000455372.2 |
SLC39A3
|
solute carrier family 39 (zinc transporter), member 3 |
| chr16_+_67562702 | 0.09 |
ENST00000379312.3
ENST00000042381.4 ENST00000540839.3 |
FAM65A
|
family with sequence similarity 65, member A |
| chr12_-_123450986 | 0.09 |
ENST00000344275.7
ENST00000442833.2 ENST00000280560.8 ENST00000540285.1 ENST00000346530.5 |
ABCB9
|
ATP-binding cassette, sub-family B (MDR/TAP), member 9 |
| chr15_+_75494214 | 0.09 |
ENST00000394987.4
|
C15orf39
|
chromosome 15 open reading frame 39 |
| chr11_+_70049269 | 0.09 |
ENST00000301838.4
|
FADD
|
Fas (TNFRSF6)-associated via death domain |
| chr10_-_126432619 | 0.09 |
ENST00000337318.3
|
FAM53B
|
family with sequence similarity 53, member B |
| chr16_+_19125252 | 0.09 |
ENST00000566735.1
ENST00000381440.3 |
ITPRIPL2
|
inositol 1,4,5-trisphosphate receptor interacting protein-like 2 |
| chr16_+_2076869 | 0.09 |
ENST00000424542.2
ENST00000432365.2 |
SLC9A3R2
|
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 2 |
| chr19_+_13051206 | 0.09 |
ENST00000586760.1
|
CALR
|
calreticulin |
| chr1_-_149908217 | 0.08 |
ENST00000369140.3
|
MTMR11
|
myotubularin related protein 11 |
| chr16_+_50313426 | 0.08 |
ENST00000569265.1
|
ADCY7
|
adenylate cyclase 7 |
| chr6_+_30844192 | 0.08 |
ENST00000502955.1
ENST00000505066.1 |
DDR1
|
discoidin domain receptor tyrosine kinase 1 |
| chr11_+_72975524 | 0.08 |
ENST00000540342.1
ENST00000542092.1 |
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr20_-_62199427 | 0.08 |
ENST00000427522.2
|
HELZ2
|
helicase with zinc finger 2, transcriptional coactivator |
| chr20_+_43343476 | 0.08 |
ENST00000372868.2
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr12_+_113416340 | 0.08 |
ENST00000552756.1
|
OAS2
|
2'-5'-oligoadenylate synthetase 2, 69/71kDa |
| chr16_-_4665023 | 0.07 |
ENST00000591897.1
|
UBALD1
|
UBA-like domain containing 1 |
| chr2_+_74120094 | 0.07 |
ENST00000409731.3
ENST00000345517.3 ENST00000409918.1 ENST00000442912.1 ENST00000409624.1 |
ACTG2
|
actin, gamma 2, smooth muscle, enteric |
| chr10_-_126432821 | 0.07 |
ENST00000280780.6
|
FAM53B
|
family with sequence similarity 53, member B |
| chr5_+_149980622 | 0.07 |
ENST00000394243.1
|
SYNPO
|
synaptopodin |
| chr5_+_137774706 | 0.07 |
ENST00000378339.2
ENST00000254901.5 ENST00000506158.1 |
REEP2
|
receptor accessory protein 2 |
| chr15_-_78112553 | 0.07 |
ENST00000562933.1
|
LINGO1
|
leucine rich repeat and Ig domain containing 1 |
| chr1_-_155224751 | 0.07 |
ENST00000350210.2
ENST00000368368.3 |
FAM189B
|
family with sequence similarity 189, member B |
| chr17_+_17685422 | 0.07 |
ENST00000395774.1
|
RAI1
|
retinoic acid induced 1 |
| chr2_+_71127699 | 0.07 |
ENST00000234392.2
|
VAX2
|
ventral anterior homeobox 2 |
| chr4_-_176733897 | 0.06 |
ENST00000393658.2
|
GPM6A
|
glycoprotein M6A |
| chr14_+_38033252 | 0.06 |
ENST00000554829.1
|
RP11-356O9.1
|
RP11-356O9.1 |
| chr1_+_11333245 | 0.06 |
ENST00000376810.5
|
UBIAD1
|
UbiA prenyltransferase domain containing 1 |
| chr15_+_40731920 | 0.06 |
ENST00000561234.1
|
BAHD1
|
bromo adjacent homology domain containing 1 |
| chr19_+_10196781 | 0.06 |
ENST00000253110.11
|
C19orf66
|
chromosome 19 open reading frame 66 |
| chr1_+_46668994 | 0.06 |
ENST00000371980.3
|
LURAP1
|
leucine rich adaptor protein 1 |
| chr7_-_72439997 | 0.06 |
ENST00000285805.3
|
TRIM74
|
tripartite motif containing 74 |
| chr5_+_127039075 | 0.06 |
ENST00000514853.2
|
CTC-228N24.1
|
CTC-228N24.1 |
| chr11_+_72975559 | 0.06 |
ENST00000349767.2
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr11_+_64808675 | 0.06 |
ENST00000529996.1
|
SAC3D1
|
SAC3 domain containing 1 |
| chr11_-_61197480 | 0.06 |
ENST00000439958.3
ENST00000394888.4 |
CPSF7
|
cleavage and polyadenylation specific factor 7, 59kDa |
| chr2_+_231902193 | 0.06 |
ENST00000373640.4
|
C2orf72
|
chromosome 2 open reading frame 72 |
| chr17_+_75372165 | 0.06 |
ENST00000427674.2
|
SEPT9
|
septin 9 |
| chr1_-_160549235 | 0.05 |
ENST00000368054.3
ENST00000368048.3 ENST00000311224.4 ENST00000368051.3 ENST00000534968.1 |
CD84
|
CD84 molecule |
| chr11_-_58343319 | 0.05 |
ENST00000395074.2
|
LPXN
|
leupaxin |
| chr19_-_3062881 | 0.05 |
ENST00000586742.1
|
AES
|
amino-terminal enhancer of split |
| chr11_+_46366918 | 0.05 |
ENST00000528615.1
ENST00000395574.3 |
DGKZ
|
diacylglycerol kinase, zeta |
| chr7_+_73588575 | 0.05 |
ENST00000265753.8
|
EIF4H
|
eukaryotic translation initiation factor 4H |
| chr7_+_72742178 | 0.05 |
ENST00000442793.1
ENST00000413573.2 ENST00000252037.4 |
FKBP6
|
FK506 binding protein 6, 36kDa |
| chr10_+_118349920 | 0.05 |
ENST00000531984.1
|
PNLIPRP1
|
pancreatic lipase-related protein 1 |
| chr4_+_39460620 | 0.05 |
ENST00000340169.2
ENST00000261434.3 |
LIAS
|
lipoic acid synthetase |
| chr9_-_123639600 | 0.05 |
ENST00000373896.3
|
PHF19
|
PHD finger protein 19 |
| chr1_+_27648709 | 0.05 |
ENST00000608611.1
ENST00000466759.1 ENST00000464813.1 ENST00000498220.1 |
TMEM222
|
transmembrane protein 222 |
| chr17_-_73150629 | 0.05 |
ENST00000356033.4
ENST00000405458.3 ENST00000409753.3 |
HN1
|
hematological and neurological expressed 1 |
| chr12_+_9066472 | 0.05 |
ENST00000538657.1
|
PHC1
|
polyhomeotic homolog 1 (Drosophila) |
| chr17_-_73150599 | 0.04 |
ENST00000392566.2
ENST00000581874.1 |
HN1
|
hematological and neurological expressed 1 |
| chr6_-_30640811 | 0.04 |
ENST00000376442.3
|
DHX16
|
DEAH (Asp-Glu-Ala-His) box polypeptide 16 |
| chr11_+_72975578 | 0.04 |
ENST00000393592.2
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr19_-_19051993 | 0.04 |
ENST00000594794.1
ENST00000355887.6 ENST00000392351.3 ENST00000596482.1 |
HOMER3
|
homer homolog 3 (Drosophila) |
| chr20_+_43343517 | 0.04 |
ENST00000372865.4
|
WISP2
|
WNT1 inducible signaling pathway protein 2 |
| chr3_-_48542197 | 0.04 |
ENST00000442747.1
ENST00000444115.1 |
SHISA5
|
shisa family member 5 |
| chr2_-_183387430 | 0.04 |
ENST00000410103.1
|
PDE1A
|
phosphodiesterase 1A, calmodulin-dependent |
| chr17_-_15370915 | 0.04 |
ENST00000312177.6
|
CDRT4
|
CMT1A duplicated region transcript 4 |
| chrX_+_109245863 | 0.04 |
ENST00000372072.3
|
TMEM164
|
transmembrane protein 164 |
| chr22_-_38484922 | 0.04 |
ENST00000428572.1
|
BAIAP2L2
|
BAI1-associated protein 2-like 2 |
| chr7_+_73588665 | 0.04 |
ENST00000353999.6
|
EIF4H
|
eukaryotic translation initiation factor 4H |
| chr6_-_44281043 | 0.04 |
ENST00000244571.4
|
AARS2
|
alanyl-tRNA synthetase 2, mitochondrial |
| chr19_+_3178736 | 0.04 |
ENST00000246115.3
|
S1PR4
|
sphingosine-1-phosphate receptor 4 |
| chr19_+_13135790 | 0.04 |
ENST00000358552.3
|
NFIX
|
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr16_-_19729453 | 0.04 |
ENST00000564480.1
|
KNOP1
|
lysine-rich nucleolar protein 1 |
| chr6_+_157099036 | 0.04 |
ENST00000350026.5
ENST00000346085.5 ENST00000367148.1 ENST00000275248.4 |
ARID1B
|
AT rich interactive domain 1B (SWI1-like) |
| chr15_+_78370140 | 0.04 |
ENST00000409568.2
|
SH2D7
|
SH2 domain containing 7 |
| chr3_+_53528659 | 0.04 |
ENST00000350061.5
|
CACNA1D
|
calcium channel, voltage-dependent, L type, alpha 1D subunit |
| chr15_-_78369994 | 0.04 |
ENST00000300584.3
ENST00000409931.3 |
TBC1D2B
|
TBC1 domain family, member 2B |
| chr11_+_46366799 | 0.04 |
ENST00000532868.2
|
DGKZ
|
diacylglycerol kinase, zeta |
| chr10_+_106113515 | 0.03 |
ENST00000369704.3
ENST00000312902.5 |
CCDC147
|
coiled-coil domain containing 147 |
| chr1_+_207277632 | 0.03 |
ENST00000421786.1
|
C4BPA
|
complement component 4 binding protein, alpha |
| chr17_+_27071002 | 0.03 |
ENST00000262395.5
ENST00000422344.1 ENST00000444415.3 ENST00000262396.6 |
TRAF4
|
TNF receptor-associated factor 4 |
| chr12_-_49319265 | 0.03 |
ENST00000552878.1
ENST00000453172.2 |
FKBP11
|
FK506 binding protein 11, 19 kDa |
| chr2_-_200320768 | 0.03 |
ENST00000428695.1
|
SATB2
|
SATB homeobox 2 |
| chr9_-_139357413 | 0.03 |
ENST00000277537.6
|
SEC16A
|
SEC16 homolog A (S. cerevisiae) |
| chr15_+_75498739 | 0.03 |
ENST00000565074.1
|
C15orf39
|
chromosome 15 open reading frame 39 |
| chr4_+_39460689 | 0.03 |
ENST00000381846.1
|
LIAS
|
lipoic acid synthetase |
| chr5_-_134734901 | 0.03 |
ENST00000312469.4
ENST00000423969.2 |
H2AFY
|
H2A histone family, member Y |
| chr17_-_39928501 | 0.03 |
ENST00000420370.1
|
JUP
|
junction plakoglobin |
| chr13_+_76445187 | 0.03 |
ENST00000318245.4
|
C13orf45
|
chromosome 13 open reading frame 45 |
| chr4_+_39460659 | 0.03 |
ENST00000513731.1
|
LIAS
|
lipoic acid synthetase |
| chr2_+_69201705 | 0.03 |
ENST00000377938.2
|
GKN1
|
gastrokine 1 |
| chr2_+_27070964 | 0.03 |
ENST00000288699.6
|
DPYSL5
|
dihydropyrimidinase-like 5 |
| chr19_+_39138320 | 0.03 |
ENST00000424234.2
ENST00000390009.3 ENST00000589528.1 |
ACTN4
|
actinin, alpha 4 |
| chr7_+_75024903 | 0.02 |
ENST00000323819.3
ENST00000430211.1 |
TRIM73
|
tripartite motif containing 73 |
| chr19_+_39647271 | 0.02 |
ENST00000599657.1
|
PAK4
|
p21 protein (Cdc42/Rac)-activated kinase 4 |
| chr19_-_344786 | 0.02 |
ENST00000264819.4
|
MIER2
|
mesoderm induction early response 1, family member 2 |
| chr17_-_26684473 | 0.02 |
ENST00000540200.1
|
POLDIP2
|
polymerase (DNA-directed), delta interacting protein 2 |
| chr19_+_16830774 | 0.02 |
ENST00000524140.2
|
NWD1
|
NACHT and WD repeat domain containing 1 |
| chr1_-_36615065 | 0.02 |
ENST00000373166.3
ENST00000373159.1 ENST00000373162.1 |
TRAPPC3
|
trafficking protein particle complex 3 |
| chr12_+_8995832 | 0.02 |
ENST00000541459.1
|
A2ML1
|
alpha-2-macroglobulin-like 1 |
| chr9_-_101558777 | 0.02 |
ENST00000375018.1
ENST00000353234.4 |
ANKS6
|
ankyrin repeat and sterile alpha motif domain containing 6 |
| chr16_+_56899114 | 0.02 |
ENST00000566786.1
ENST00000438926.2 ENST00000563236.1 ENST00000262502.5 |
SLC12A3
|
solute carrier family 12 (sodium/chloride transporter), member 3 |
| chr17_+_5031687 | 0.02 |
ENST00000250066.6
ENST00000304328.5 |
USP6
|
ubiquitin specific peptidase 6 (Tre-2 oncogene) |
| chr1_-_27709793 | 0.02 |
ENST00000374027.3
ENST00000374025.3 |
CD164L2
|
CD164 sialomucin-like 2 |
| chr2_+_27071045 | 0.02 |
ENST00000401478.1
|
DPYSL5
|
dihydropyrimidinase-like 5 |
| chrX_-_15402498 | 0.02 |
ENST00000297904.3
|
FIGF
|
c-fos induced growth factor (vascular endothelial growth factor D) |
| chr12_+_25055243 | 0.02 |
ENST00000599478.1
|
AC026310.1
|
Protein 101060047 |
| chr2_+_27071292 | 0.01 |
ENST00000431402.1
ENST00000434719.1 |
DPYSL5
|
dihydropyrimidinase-like 5 |
| chr4_-_186732048 | 0.01 |
ENST00000448662.2
ENST00000439049.1 ENST00000420158.1 ENST00000431808.1 ENST00000319471.9 |
SORBS2
|
sorbin and SH3 domain containing 2 |
| chr2_-_158345462 | 0.01 |
ENST00000439355.1
ENST00000540637.1 |
CYTIP
|
cytohesin 1 interacting protein |
| chr16_+_8736232 | 0.01 |
ENST00000562973.1
|
METTL22
|
methyltransferase like 22 |
| chr7_+_72742162 | 0.01 |
ENST00000431982.2
|
FKBP6
|
FK506 binding protein 6, 36kDa |
| chr1_-_53704157 | 0.01 |
ENST00000371466.4
ENST00000371470.3 |
MAGOH
|
mago-nashi homolog, proliferation-associated (Drosophila) |
| chr15_+_78384901 | 0.01 |
ENST00000328828.5
|
SH2D7
|
SH2 domain containing 7 |
| chr3_-_179322436 | 0.01 |
ENST00000392659.2
ENST00000476781.1 |
MRPL47
|
mitochondrial ribosomal protein L47 |
| chr10_+_23384435 | 0.01 |
ENST00000376510.3
|
MSRB2
|
methionine sulfoxide reductase B2 |
| chr9_-_123639445 | 0.01 |
ENST00000312189.6
|
PHF19
|
PHD finger protein 19 |
| chr11_-_568369 | 0.01 |
ENST00000534540.1
ENST00000528245.1 ENST00000500447.1 ENST00000533920.1 |
MIR210HG
|
MIR210 host gene (non-protein coding) |
| chr12_-_25055177 | 0.01 |
ENST00000538118.1
|
BCAT1
|
branched chain amino-acid transaminase 1, cytosolic |
| chr14_-_67982146 | 0.01 |
ENST00000557779.1
ENST00000557006.1 |
TMEM229B
|
transmembrane protein 229B |
| chr3_+_38388251 | 0.01 |
ENST00000427323.1
ENST00000207870.3 ENST00000542835.1 |
XYLB
|
xylulokinase homolog (H. influenzae) |
| chr11_-_73720276 | 0.00 |
ENST00000348534.4
|
UCP3
|
uncoupling protein 3 (mitochondrial, proton carrier) |
| chr3_+_51663407 | 0.00 |
ENST00000432863.1
ENST00000296477.3 |
RAD54L2
|
RAD54-like 2 (S. cerevisiae) |
| chr1_-_51796987 | 0.00 |
ENST00000262676.5
|
TTC39A
|
tetratricopeptide repeat domain 39A |
| chr3_-_47023455 | 0.00 |
ENST00000446836.1
ENST00000425441.1 |
CCDC12
|
coiled-coil domain containing 12 |
| chr6_-_30640761 | 0.00 |
ENST00000415603.1
|
DHX16
|
DEAH (Asp-Glu-Ala-His) box polypeptide 16 |
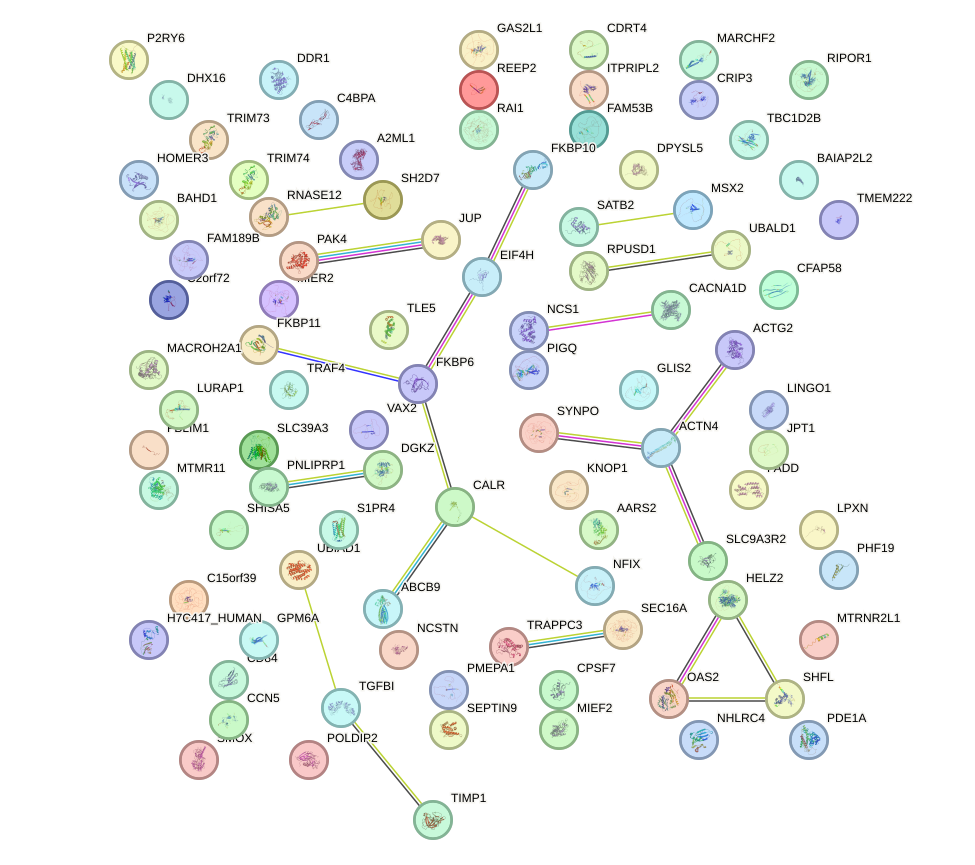
Network of associatons between targets according to the STRING database.
First level regulatory network of ZKSCAN1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0051795 | positive regulation of catagen(GO:0051795) |
| 0.0 | 0.3 | GO:0060994 | regulation of transcription from RNA polymerase II promoter involved in kidney development(GO:0060994) |
| 0.0 | 0.3 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.1 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.0 | 0.1 | GO:0009107 | lipoate biosynthetic process(GO:0009107) |
| 0.0 | 0.3 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.1 | GO:0002396 | MHC protein complex assembly(GO:0002396) peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.0 | 0.2 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.1 | GO:0035672 | oligopeptide transmembrane transport(GO:0035672) |
| 0.0 | 0.1 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.0 | 0.2 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.0 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.1 | GO:0097527 | necroptotic signaling pathway(GO:0097527) |
| 0.0 | 0.1 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.1 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.1 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) |
| 0.0 | 0.1 | GO:0009233 | menaquinone metabolic process(GO:0009233) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.0 | 0.1 | GO:0070765 | gamma-secretase complex(GO:0070765) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0016992 | lipoate synthase activity(GO:0016992) radical SAM enzyme activity(GO:0070283) |
| 0.0 | 0.1 | GO:0052895 | norspermine:oxygen oxidoreductase activity(GO:0052894) N1-acetylspermine:oxygen oxidoreductase (N1-acetylspermidine-forming) activity(GO:0052895) |
| 0.0 | 0.2 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 0.1 | GO:0034057 | RNA strand-exchange activity(GO:0034057) |
| 0.0 | 0.1 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.0 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |