Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
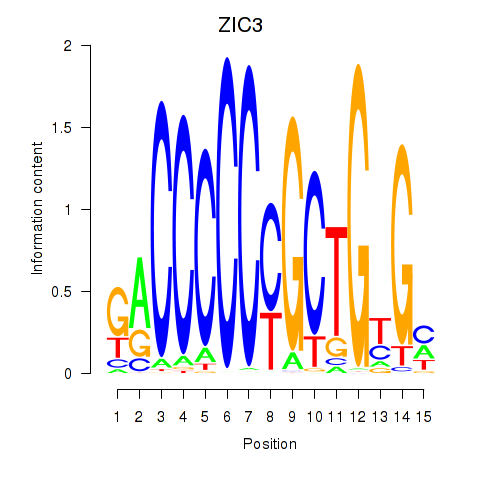
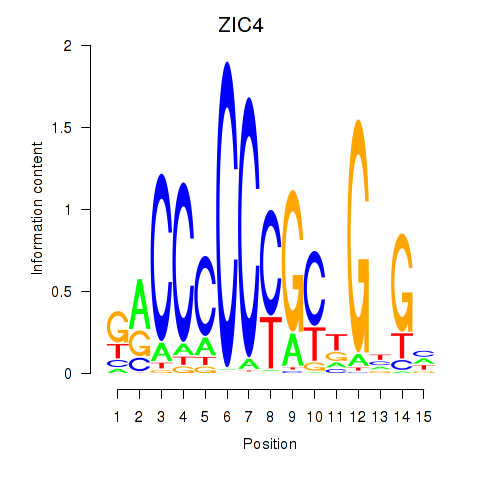
Results for ZIC3_ZIC4
Z-value: 0.57
Transcription factors associated with ZIC3_ZIC4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZIC3
|
ENSG00000156925.7 | Zic family member 3 |
|
ZIC4
|
ENSG00000174963.13 | Zic family member 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZIC4 | hg19_v2_chr3_-_147124547_147124647 | -0.43 | 5.7e-01 | Click! |
Activity profile of ZIC3_ZIC4 motif
Sorted Z-values of ZIC3_ZIC4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_302918 | 0.34 |
ENST00000599994.1
|
AC187652.1
|
Protein LOC100996433 |
| chr1_+_38273988 | 0.22 |
ENST00000446260.2
|
C1orf122
|
chromosome 1 open reading frame 122 |
| chr1_+_38273818 | 0.22 |
ENST00000373042.4
|
C1orf122
|
chromosome 1 open reading frame 122 |
| chr9_+_19408919 | 0.20 |
ENST00000380376.1
|
ACER2
|
alkaline ceramidase 2 |
| chr11_-_67272794 | 0.15 |
ENST00000436757.2
ENST00000356404.3 |
PITPNM1
|
phosphatidylinositol transfer protein, membrane-associated 1 |
| chr17_-_78194124 | 0.15 |
ENST00000570427.1
ENST00000570923.1 |
SGSH
|
N-sulfoglucosamine sulfohydrolase |
| chr10_+_71561630 | 0.15 |
ENST00000398974.3
ENST00000398971.3 ENST00000398968.3 ENST00000398966.3 ENST00000398964.3 ENST00000398969.3 ENST00000356340.3 ENST00000398972.3 ENST00000398973.3 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr17_-_77179487 | 0.14 |
ENST00000580508.1
|
RBFOX3
|
RNA binding protein, fox-1 homolog (C. elegans) 3 |
| chr12_+_132312931 | 0.14 |
ENST00000360564.1
ENST00000545671.1 ENST00000545790.1 |
MMP17
|
matrix metallopeptidase 17 (membrane-inserted) |
| chr2_+_105050794 | 0.14 |
ENST00000429464.1
ENST00000414442.1 ENST00000447380.1 |
AC013402.2
|
long intergenic non-protein coding RNA 1102 |
| chr3_+_143690640 | 0.13 |
ENST00000315691.3
|
C3orf58
|
chromosome 3 open reading frame 58 |
| chrX_+_2747306 | 0.13 |
ENST00000520904.1
|
GYG2
|
glycogenin 2 |
| chr1_-_32801825 | 0.13 |
ENST00000329421.7
|
MARCKSL1
|
MARCKS-like 1 |
| chr19_+_30156400 | 0.13 |
ENST00000588833.1
|
PLEKHF1
|
pleckstrin homology domain containing, family F (with FYVE domain) member 1 |
| chr18_+_2846972 | 0.13 |
ENST00000254528.3
|
EMILIN2
|
elastin microfibril interfacer 2 |
| chr17_-_40828969 | 0.12 |
ENST00000591022.1
ENST00000587627.1 ENST00000293349.6 |
PLEKHH3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr16_-_88717482 | 0.12 |
ENST00000261623.3
|
CYBA
|
cytochrome b-245, alpha polypeptide |
| chr19_+_1908257 | 0.12 |
ENST00000411971.1
|
SCAMP4
|
secretory carrier membrane protein 4 |
| chr3_-_134093738 | 0.11 |
ENST00000506107.1
|
AMOTL2
|
angiomotin like 2 |
| chr3_-_64253655 | 0.11 |
ENST00000498162.1
|
PRICKLE2
|
prickle homolog 2 (Drosophila) |
| chr2_+_219264762 | 0.11 |
ENST00000452977.1
|
CTDSP1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1 |
| chr11_+_66790816 | 0.11 |
ENST00000527043.1
|
SYT12
|
synaptotagmin XII |
| chr19_-_40724246 | 0.10 |
ENST00000311308.6
|
TTC9B
|
tetratricopeptide repeat domain 9B |
| chr7_-_994302 | 0.10 |
ENST00000265846.5
|
ADAP1
|
ArfGAP with dual PH domains 1 |
| chr7_+_65540780 | 0.10 |
ENST00000304874.9
|
ASL
|
argininosuccinate lyase |
| chr17_-_40829026 | 0.10 |
ENST00000412503.1
|
PLEKHH3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr7_+_65540853 | 0.10 |
ENST00000380839.4
ENST00000395332.3 ENST00000362000.5 ENST00000395331.3 |
ASL
|
argininosuccinate lyase |
| chr12_-_48213735 | 0.10 |
ENST00000417902.1
ENST00000417107.1 |
HDAC7
|
histone deacetylase 7 |
| chr15_-_91565743 | 0.10 |
ENST00000535843.1
|
VPS33B
|
vacuolar protein sorting 33 homolog B (yeast) |
| chr3_+_71803201 | 0.09 |
ENST00000304411.2
|
GPR27
|
G protein-coupled receptor 27 |
| chr10_-_79397547 | 0.09 |
ENST00000481070.1
|
KCNMA1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr15_-_91565770 | 0.09 |
ENST00000535906.1
ENST00000333371.3 |
VPS33B
|
vacuolar protein sorting 33 homolog B (yeast) |
| chr1_-_16400086 | 0.09 |
ENST00000375662.4
|
FAM131C
|
family with sequence similarity 131, member C |
| chr17_-_73840415 | 0.09 |
ENST00000592386.1
ENST00000412096.2 ENST00000586147.1 |
UNC13D
|
unc-13 homolog D (C. elegans) |
| chr10_-_9801179 | 0.09 |
ENST00000419836.1
|
RP5-1051H14.2
|
RP5-1051H14.2 |
| chr14_+_57857262 | 0.09 |
ENST00000555166.1
ENST00000556492.1 ENST00000554703.1 |
NAA30
|
N(alpha)-acetyltransferase 30, NatC catalytic subunit |
| chr16_+_2570340 | 0.08 |
ENST00000568263.1
ENST00000293971.6 ENST00000302956.4 ENST00000413459.3 ENST00000566706.1 ENST00000569879.1 |
AMDHD2
|
amidohydrolase domain containing 2 |
| chr1_+_6673745 | 0.08 |
ENST00000377648.4
|
PHF13
|
PHD finger protein 13 |
| chr5_+_150406527 | 0.08 |
ENST00000520059.1
|
GPX3
|
glutathione peroxidase 3 (plasma) |
| chr5_+_421030 | 0.08 |
ENST00000506456.1
|
AHRR
|
aryl-hydrocarbon receptor repressor |
| chr5_+_176873789 | 0.08 |
ENST00000323249.3
ENST00000502922.1 |
PRR7
|
proline rich 7 (synaptic) |
| chr11_-_67276100 | 0.08 |
ENST00000301488.3
|
CDK2AP2
|
cyclin-dependent kinase 2 associated protein 2 |
| chr8_-_90769939 | 0.08 |
ENST00000504145.1
ENST00000519655.2 |
RP11-37B2.1
|
RP11-37B2.1 |
| chr19_+_47759716 | 0.08 |
ENST00000221922.6
|
CCDC9
|
coiled-coil domain containing 9 |
| chr9_+_139738867 | 0.08 |
ENST00000436881.1
|
C9orf172
|
chromosome 9 open reading frame 172 |
| chr22_+_29702572 | 0.08 |
ENST00000407647.2
ENST00000416823.1 ENST00000428622.1 |
GAS2L1
|
growth arrest-specific 2 like 1 |
| chr7_-_92107229 | 0.08 |
ENST00000603053.1
|
ERVW-1
|
endogenous retrovirus group W, member 1 |
| chr19_-_40791211 | 0.08 |
ENST00000579047.1
|
AKT2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr14_+_100848311 | 0.08 |
ENST00000542471.2
|
WDR25
|
WD repeat domain 25 |
| chr11_-_407103 | 0.08 |
ENST00000526395.1
|
SIGIRR
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chr3_-_129035120 | 0.07 |
ENST00000333762.4
|
H1FX
|
H1 histone family, member X |
| chr10_+_71561649 | 0.07 |
ENST00000398978.3
ENST00000354547.3 ENST00000357811.3 |
COL13A1
|
collagen, type XIII, alpha 1 |
| chr9_-_130742792 | 0.07 |
ENST00000373095.1
|
FAM102A
|
family with sequence similarity 102, member A |
| chr11_-_62477313 | 0.07 |
ENST00000464544.1
ENST00000530009.1 |
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr20_+_61340179 | 0.07 |
ENST00000370501.3
|
NTSR1
|
neurotensin receptor 1 (high affinity) |
| chr7_-_5570229 | 0.07 |
ENST00000331789.5
|
ACTB
|
actin, beta |
| chr19_+_54695098 | 0.07 |
ENST00000396388.2
|
TSEN34
|
TSEN34 tRNA splicing endonuclease subunit |
| chr1_-_38273840 | 0.07 |
ENST00000373044.2
|
YRDC
|
yrdC N(6)-threonylcarbamoyltransferase domain containing |
| chr1_-_6240183 | 0.07 |
ENST00000262450.3
ENST00000378021.1 |
CHD5
|
chromodomain helicase DNA binding protein 5 |
| chr12_+_58176525 | 0.07 |
ENST00000543727.1
ENST00000540550.1 ENST00000323833.8 ENST00000350762.5 ENST00000550559.1 ENST00000548851.1 ENST00000434359.1 ENST00000457189.1 |
TSFM
|
Ts translation elongation factor, mitochondrial |
| chr12_+_49932886 | 0.07 |
ENST00000257981.6
|
KCNH3
|
potassium voltage-gated channel, subfamily H (eag-related), member 3 |
| chr1_-_45308616 | 0.07 |
ENST00000447098.2
ENST00000372192.3 |
PTCH2
|
patched 2 |
| chr17_-_4463856 | 0.07 |
ENST00000574584.1
ENST00000381550.3 ENST00000301395.3 |
GGT6
|
gamma-glutamyltransferase 6 |
| chr19_+_30156351 | 0.06 |
ENST00000592810.1
|
PLEKHF1
|
pleckstrin homology domain containing, family F (with FYVE domain) member 1 |
| chr1_+_33352036 | 0.06 |
ENST00000373467.3
|
HPCA
|
hippocalcin |
| chr17_+_45331184 | 0.06 |
ENST00000559488.1
ENST00000571680.1 ENST00000435993.2 |
ITGB3
|
integrin, beta 3 (platelet glycoprotein IIIa, antigen CD61) |
| chr19_+_49122548 | 0.06 |
ENST00000245222.4
ENST00000340932.3 ENST00000601712.1 ENST00000600537.1 |
SPHK2
|
sphingosine kinase 2 |
| chr6_+_15249128 | 0.06 |
ENST00000397311.3
|
JARID2
|
jumonji, AT rich interactive domain 2 |
| chr1_+_180875711 | 0.06 |
ENST00000434447.1
|
RP11-46A10.2
|
RP11-46A10.2 |
| chr5_+_176873446 | 0.06 |
ENST00000507881.1
|
PRR7
|
proline rich 7 (synaptic) |
| chr2_-_98280383 | 0.06 |
ENST00000289228.5
|
ACTR1B
|
ARP1 actin-related protein 1 homolog B, centractin beta (yeast) |
| chr10_+_134145735 | 0.06 |
ENST00000368613.4
|
LRRC27
|
leucine rich repeat containing 27 |
| chr6_-_32145861 | 0.06 |
ENST00000336984.6
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr6_-_38607673 | 0.06 |
ENST00000481247.1
|
BTBD9
|
BTB (POZ) domain containing 9 |
| chr17_-_78194716 | 0.06 |
ENST00000576707.1
|
SGSH
|
N-sulfoglucosamine sulfohydrolase |
| chr7_-_5569588 | 0.06 |
ENST00000417101.1
|
ACTB
|
actin, beta |
| chr1_-_212873267 | 0.06 |
ENST00000243440.1
|
BATF3
|
basic leucine zipper transcription factor, ATF-like 3 |
| chr17_+_77030267 | 0.06 |
ENST00000581774.1
|
C1QTNF1
|
C1q and tumor necrosis factor related protein 1 |
| chr10_+_45455207 | 0.06 |
ENST00000334940.6
ENST00000374417.2 ENST00000340258.5 ENST00000427758.1 |
RASSF4
|
Ras association (RalGDS/AF-6) domain family member 4 |
| chr16_+_9185450 | 0.06 |
ENST00000327827.7
|
C16orf72
|
chromosome 16 open reading frame 72 |
| chr19_-_51587502 | 0.06 |
ENST00000156499.2
ENST00000391802.1 |
KLK14
|
kallikrein-related peptidase 14 |
| chr7_+_150758304 | 0.06 |
ENST00000482950.1
ENST00000463414.1 ENST00000310317.5 |
SLC4A2
|
solute carrier family 4 (anion exchanger), member 2 |
| chr11_-_2906979 | 0.06 |
ENST00000380725.1
ENST00000313407.6 ENST00000430149.2 ENST00000440480.2 ENST00000414822.3 |
CDKN1C
|
cyclin-dependent kinase inhibitor 1C (p57, Kip2) |
| chr22_-_26986045 | 0.06 |
ENST00000442495.1
ENST00000440953.1 ENST00000450022.1 ENST00000338754.4 |
TPST2
|
tyrosylprotein sulfotransferase 2 |
| chr19_+_13051206 | 0.06 |
ENST00000586760.1
|
CALR
|
calreticulin |
| chr19_-_12992244 | 0.06 |
ENST00000538460.1
|
DNASE2
|
deoxyribonuclease II, lysosomal |
| chr19_+_45251804 | 0.06 |
ENST00000164227.5
|
BCL3
|
B-cell CLL/lymphoma 3 |
| chr16_+_777739 | 0.06 |
ENST00000563792.1
|
HAGHL
|
hydroxyacylglutathione hydrolase-like |
| chr19_-_40791160 | 0.06 |
ENST00000358335.5
|
AKT2
|
v-akt murine thymoma viral oncogene homolog 2 |
| chr17_-_37844267 | 0.06 |
ENST00000579146.1
ENST00000378011.4 ENST00000429199.2 ENST00000300658.4 |
PGAP3
|
post-GPI attachment to proteins 3 |
| chr19_+_10362882 | 0.06 |
ENST00000393733.2
ENST00000588502.1 |
MRPL4
|
mitochondrial ribosomal protein L4 |
| chr19_+_33685490 | 0.06 |
ENST00000253193.7
|
LRP3
|
low density lipoprotein receptor-related protein 3 |
| chr1_-_52521831 | 0.05 |
ENST00000371626.4
|
TXNDC12
|
thioredoxin domain containing 12 (endoplasmic reticulum) |
| chr19_+_48949030 | 0.05 |
ENST00000253237.5
|
GRWD1
|
glutamate-rich WD repeat containing 1 |
| chr11_-_65150103 | 0.05 |
ENST00000294187.6
ENST00000398802.1 ENST00000360662.3 ENST00000377152.2 ENST00000530936.1 |
SLC25A45
|
solute carrier family 25, member 45 |
| chr7_+_74379083 | 0.05 |
ENST00000361825.7
|
GATSL1
|
GATS protein-like 1 |
| chr16_+_58035277 | 0.05 |
ENST00000219281.3
ENST00000539737.2 ENST00000423271.3 ENST00000561568.1 ENST00000563149.1 |
USB1
|
U6 snRNA biogenesis 1 |
| chr5_-_176923846 | 0.05 |
ENST00000506537.1
|
PDLIM7
|
PDZ and LIM domain 7 (enigma) |
| chrX_-_19533379 | 0.05 |
ENST00000338883.4
|
MAP3K15
|
mitogen-activated protein kinase kinase kinase 15 |
| chr5_+_176731572 | 0.05 |
ENST00000503853.1
|
PRELID1
|
PRELI domain containing 1 |
| chr3_+_187420101 | 0.05 |
ENST00000449623.1
ENST00000437407.1 |
RP11-211G3.3
|
Uncharacterized protein |
| chr22_-_41842781 | 0.05 |
ENST00000434408.1
|
TOB2
|
transducer of ERBB2, 2 |
| chr17_+_39421591 | 0.05 |
ENST00000391355.1
|
KRTAP9-6
|
keratin associated protein 9-6 |
| chr20_+_35201857 | 0.05 |
ENST00000373874.2
|
TGIF2
|
TGFB-induced factor homeobox 2 |
| chr2_+_219125714 | 0.05 |
ENST00000522678.1
ENST00000519574.1 ENST00000521462.1 |
GPBAR1
|
G protein-coupled bile acid receptor 1 |
| chr19_+_48949087 | 0.05 |
ENST00000598711.1
|
GRWD1
|
glutamate-rich WD repeat containing 1 |
| chr15_+_75074385 | 0.05 |
ENST00000220003.9
|
CSK
|
c-src tyrosine kinase |
| chr19_+_49617609 | 0.05 |
ENST00000221459.2
ENST00000486217.2 |
LIN7B
|
lin-7 homolog B (C. elegans) |
| chr19_-_49314269 | 0.05 |
ENST00000545387.2
ENST00000316273.6 ENST00000402551.1 ENST00000598162.1 ENST00000599246.1 |
BCAT2
|
branched chain amino-acid transaminase 2, mitochondrial |
| chr20_+_49411523 | 0.05 |
ENST00000371608.2
|
BCAS4
|
breast carcinoma amplified sequence 4 |
| chr1_+_52521797 | 0.05 |
ENST00000313334.8
|
BTF3L4
|
basic transcription factor 3-like 4 |
| chr19_+_59055814 | 0.05 |
ENST00000594806.1
ENST00000253024.5 ENST00000341753.6 |
TRIM28
|
tripartite motif containing 28 |
| chr9_-_140082983 | 0.05 |
ENST00000323927.2
|
ANAPC2
|
anaphase promoting complex subunit 2 |
| chr8_+_22250334 | 0.05 |
ENST00000520832.1
|
SLC39A14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr17_+_78194261 | 0.05 |
ENST00000572725.1
|
SLC26A11
|
solute carrier family 26 (anion exchanger), member 11 |
| chr17_+_260097 | 0.05 |
ENST00000360127.6
ENST00000571106.1 ENST00000491373.1 |
C17orf97
|
chromosome 17 open reading frame 97 |
| chr19_-_49122384 | 0.05 |
ENST00000550973.1
ENST00000550645.1 ENST00000549273.1 ENST00000552588.1 |
RPL18
|
ribosomal protein L18 |
| chr11_+_134146635 | 0.05 |
ENST00000431683.2
|
GLB1L3
|
galactosidase, beta 1-like 3 |
| chr16_+_29466426 | 0.05 |
ENST00000567248.1
|
SLX1B
|
SLX1 structure-specific endonuclease subunit homolog B (S. cerevisiae) |
| chr17_+_78194205 | 0.05 |
ENST00000573809.1
ENST00000361193.3 ENST00000574967.1 ENST00000576126.1 ENST00000411502.3 ENST00000546047.2 |
SLC26A11
|
solute carrier family 26 (anion exchanger), member 11 |
| chr16_+_30205225 | 0.05 |
ENST00000345535.4
ENST00000251303.6 |
SLX1A
|
SLX1 structure-specific endonuclease subunit homolog A (S. cerevisiae) |
| chr19_+_45409011 | 0.05 |
ENST00000252486.4
ENST00000446996.1 ENST00000434152.1 |
APOE
|
apolipoprotein E |
| chr11_-_73694346 | 0.05 |
ENST00000310473.3
|
UCP2
|
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr9_-_123639445 | 0.05 |
ENST00000312189.6
|
PHF19
|
PHD finger protein 19 |
| chr1_+_28995231 | 0.05 |
ENST00000373816.1
|
GMEB1
|
glucocorticoid modulatory element binding protein 1 |
| chr4_-_860950 | 0.04 |
ENST00000511980.1
ENST00000510799.1 |
GAK
|
cyclin G associated kinase |
| chr11_-_125932685 | 0.04 |
ENST00000527967.1
|
CDON
|
cell adhesion associated, oncogene regulated |
| chr6_+_30882108 | 0.04 |
ENST00000541562.1
ENST00000421263.1 |
VARS2
|
valyl-tRNA synthetase 2, mitochondrial |
| chr11_+_73019282 | 0.04 |
ENST00000263674.3
|
ARHGEF17
|
Rho guanine nucleotide exchange factor (GEF) 17 |
| chr17_+_45771420 | 0.04 |
ENST00000578982.1
|
TBKBP1
|
TBK1 binding protein 1 |
| chr9_+_134269439 | 0.04 |
ENST00000405995.1
|
PRRC2B
|
proline-rich coiled-coil 2B |
| chr10_+_112257596 | 0.04 |
ENST00000369583.3
|
DUSP5
|
dual specificity phosphatase 5 |
| chr14_+_24630465 | 0.04 |
ENST00000557894.1
ENST00000559284.1 ENST00000560275.1 |
IRF9
|
interferon regulatory factor 9 |
| chr21_+_46825032 | 0.04 |
ENST00000400337.2
|
COL18A1
|
collagen, type XVIII, alpha 1 |
| chr5_+_132202252 | 0.04 |
ENST00000378670.3
ENST00000378667.1 ENST00000378665.1 |
UQCRQ
|
ubiquinol-cytochrome c reductase, complex III subunit VII, 9.5kDa |
| chr1_-_6052463 | 0.04 |
ENST00000378156.4
|
NPHP4
|
nephronophthisis 4 |
| chr10_+_134145614 | 0.04 |
ENST00000368615.3
ENST00000392638.2 ENST00000344079.5 ENST00000356571.4 ENST00000368614.3 |
LRRC27
|
leucine rich repeat containing 27 |
| chr16_+_30205754 | 0.04 |
ENST00000354723.6
ENST00000355544.5 |
SULT1A3
|
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 |
| chr2_+_20646824 | 0.04 |
ENST00000272233.4
|
RHOB
|
ras homolog family member B |
| chr17_+_39411636 | 0.04 |
ENST00000394008.1
|
KRTAP9-9
|
keratin associated protein 9-9 |
| chr7_+_75511362 | 0.04 |
ENST00000428119.1
|
RHBDD2
|
rhomboid domain containing 2 |
| chr16_+_89989687 | 0.04 |
ENST00000315491.7
ENST00000555576.1 ENST00000554336.1 ENST00000553967.1 |
TUBB3
|
Tubulin beta-3 chain |
| chrX_+_153686614 | 0.04 |
ENST00000369682.3
|
PLXNA3
|
plexin A3 |
| chr15_+_75074410 | 0.04 |
ENST00000439220.2
|
CSK
|
c-src tyrosine kinase |
| chr2_-_42721110 | 0.04 |
ENST00000394973.4
ENST00000306078.1 |
KCNG3
|
potassium voltage-gated channel, subfamily G, member 3 |
| chr19_-_1863567 | 0.04 |
ENST00000250916.4
|
KLF16
|
Kruppel-like factor 16 |
| chr11_-_62477041 | 0.04 |
ENST00000433053.1
|
BSCL2
|
Berardinelli-Seip congenital lipodystrophy 2 (seipin) |
| chr16_+_2570431 | 0.04 |
ENST00000563556.1
|
AMDHD2
|
amidohydrolase domain containing 2 |
| chr16_-_2059797 | 0.04 |
ENST00000563630.1
|
ZNF598
|
zinc finger protein 598 |
| chr14_+_96342729 | 0.04 |
ENST00000504119.1
|
LINC00617
|
long intergenic non-protein coding RNA 617 |
| chr1_-_159915386 | 0.04 |
ENST00000361509.3
ENST00000368094.1 |
IGSF9
|
immunoglobulin superfamily, member 9 |
| chr4_+_2819883 | 0.04 |
ENST00000511747.1
ENST00000503393.2 |
SH3BP2
|
SH3-domain binding protein 2 |
| chr12_-_51420108 | 0.04 |
ENST00000547198.1
|
SLC11A2
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr17_-_74582191 | 0.04 |
ENST00000225276.5
|
ST6GALNAC2
|
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 2 |
| chr7_+_120969045 | 0.04 |
ENST00000222462.2
|
WNT16
|
wingless-type MMTV integration site family, member 16 |
| chr17_+_47572647 | 0.04 |
ENST00000172229.3
|
NGFR
|
nerve growth factor receptor |
| chr10_-_134145321 | 0.04 |
ENST00000368625.4
ENST00000368619.3 ENST00000456004.1 ENST00000368620.2 |
STK32C
|
serine/threonine kinase 32C |
| chr3_+_50606577 | 0.04 |
ENST00000434410.1
ENST00000232854.4 |
HEMK1
|
HemK methyltransferase family member 1 |
| chr9_+_140083099 | 0.04 |
ENST00000322310.5
|
SSNA1
|
Sjogren syndrome nuclear autoantigen 1 |
| chrX_+_152783131 | 0.04 |
ENST00000349466.2
ENST00000370186.1 |
ATP2B3
|
ATPase, Ca++ transporting, plasma membrane 3 |
| chr22_-_19165917 | 0.04 |
ENST00000451283.1
|
SLC25A1
|
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1 |
| chr1_-_25256368 | 0.04 |
ENST00000308873.6
|
RUNX3
|
runt-related transcription factor 3 |
| chrX_+_71996972 | 0.04 |
ENST00000334036.5
|
DMRTC1B
|
DMRT-like family C1B |
| chr22_-_19466732 | 0.04 |
ENST00000263202.10
ENST00000360834.4 |
UFD1L
|
ubiquitin fusion degradation 1 like (yeast) |
| chr19_-_12833361 | 0.04 |
ENST00000592287.1
|
TNPO2
|
transportin 2 |
| chr20_-_32274179 | 0.04 |
ENST00000343380.5
|
E2F1
|
E2F transcription factor 1 |
| chr2_-_96811170 | 0.04 |
ENST00000288943.4
|
DUSP2
|
dual specificity phosphatase 2 |
| chr19_-_45735138 | 0.04 |
ENST00000252482.3
|
EXOC3L2
|
exocyst complex component 3-like 2 |
| chr1_+_52521928 | 0.04 |
ENST00000489308.2
|
BTF3L4
|
basic transcription factor 3-like 4 |
| chr14_-_23822080 | 0.04 |
ENST00000397267.1
ENST00000354772.3 |
SLC22A17
|
solute carrier family 22, member 17 |
| chr2_-_10220538 | 0.04 |
ENST00000381813.4
|
CYS1
|
cystin 1 |
| chr1_-_234745234 | 0.04 |
ENST00000366610.3
ENST00000366609.3 |
IRF2BP2
|
interferon regulatory factor 2 binding protein 2 |
| chr17_+_38599693 | 0.04 |
ENST00000542955.1
ENST00000269593.4 |
IGFBP4
|
insulin-like growth factor binding protein 4 |
| chr6_+_30881982 | 0.04 |
ENST00000321897.5
ENST00000416670.2 ENST00000542001.1 ENST00000428017.1 |
VARS2
|
valyl-tRNA synthetase 2, mitochondrial |
| chr3_-_9811595 | 0.04 |
ENST00000256460.3
|
CAMK1
|
calcium/calmodulin-dependent protein kinase I |
| chr12_+_121163538 | 0.04 |
ENST00000242592.4
|
ACADS
|
acyl-CoA dehydrogenase, C-2 to C-3 short chain |
| chr19_+_17970693 | 0.04 |
ENST00000600147.1
ENST00000599898.1 |
RPL18A
|
ribosomal protein L18a |
| chr14_+_64971438 | 0.04 |
ENST00000555321.1
|
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr11_+_120081475 | 0.04 |
ENST00000328965.4
|
OAF
|
OAF homolog (Drosophila) |
| chr5_+_139055021 | 0.04 |
ENST00000502716.1
ENST00000503511.1 |
CXXC5
|
CXXC finger protein 5 |
| chr22_-_50683381 | 0.04 |
ENST00000439308.2
|
TUBGCP6
|
tubulin, gamma complex associated protein 6 |
| chr9_-_140095186 | 0.03 |
ENST00000409012.4
|
TPRN
|
taperin |
| chr1_+_156024525 | 0.03 |
ENST00000368305.4
|
LAMTOR2
|
late endosomal/lysosomal adaptor, MAPK and MTOR activator 2 |
| chr3_-_49203744 | 0.03 |
ENST00000321895.6
|
CCDC71
|
coiled-coil domain containing 71 |
| chr17_-_1083078 | 0.03 |
ENST00000574266.1
ENST00000302538.5 |
ABR
|
active BCR-related |
| chr4_-_15656923 | 0.03 |
ENST00000382358.4
ENST00000412094.2 ENST00000341285.3 |
FBXL5
|
F-box and leucine-rich repeat protein 5 |
| chr11_-_2160180 | 0.03 |
ENST00000381406.4
|
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
| chr2_+_234637754 | 0.03 |
ENST00000482026.1
ENST00000609767.1 |
UGT1A3
UGT1A1
|
UDP glucuronosyltransferase 1 family, polypeptide A3 UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr2_+_65216462 | 0.03 |
ENST00000234256.3
|
SLC1A4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr5_+_76506706 | 0.03 |
ENST00000340978.3
ENST00000346042.3 ENST00000264917.5 ENST00000342343.4 ENST00000333194.4 |
PDE8B
|
phosphodiesterase 8B |
| chr16_+_58059470 | 0.03 |
ENST00000219271.3
|
MMP15
|
matrix metallopeptidase 15 (membrane-inserted) |
| chr19_-_53193731 | 0.03 |
ENST00000598536.1
ENST00000594682.2 ENST00000601257.1 |
ZNF83
|
zinc finger protein 83 |
| chr2_-_197791441 | 0.03 |
ENST00000409475.1
ENST00000354764.4 ENST00000374738.3 |
PGAP1
|
post-GPI attachment to proteins 1 |
| chr16_+_30996502 | 0.03 |
ENST00000353250.5
ENST00000262520.6 ENST00000297679.5 ENST00000562932.1 ENST00000574447.1 |
HSD3B7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr1_+_3370990 | 0.03 |
ENST00000378378.4
|
ARHGEF16
|
Rho guanine nucleotide exchange factor (GEF) 16 |
| chr16_+_29818857 | 0.03 |
ENST00000567444.1
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr19_+_17858509 | 0.03 |
ENST00000594202.1
ENST00000252771.7 ENST00000389133.4 |
FCHO1
|
FCH domain only 1 |
| chr16_+_29465822 | 0.03 |
ENST00000330181.5
ENST00000351581.4 |
SLX1B
|
SLX1 structure-specific endonuclease subunit homolog B (S. cerevisiae) |
| chr11_+_101981423 | 0.03 |
ENST00000531439.1
|
YAP1
|
Yes-associated protein 1 |
| chr11_-_2158507 | 0.03 |
ENST00000381392.1
ENST00000381395.1 ENST00000418738.2 |
IGF2
|
insulin-like growth factor 2 (somatomedin A) |
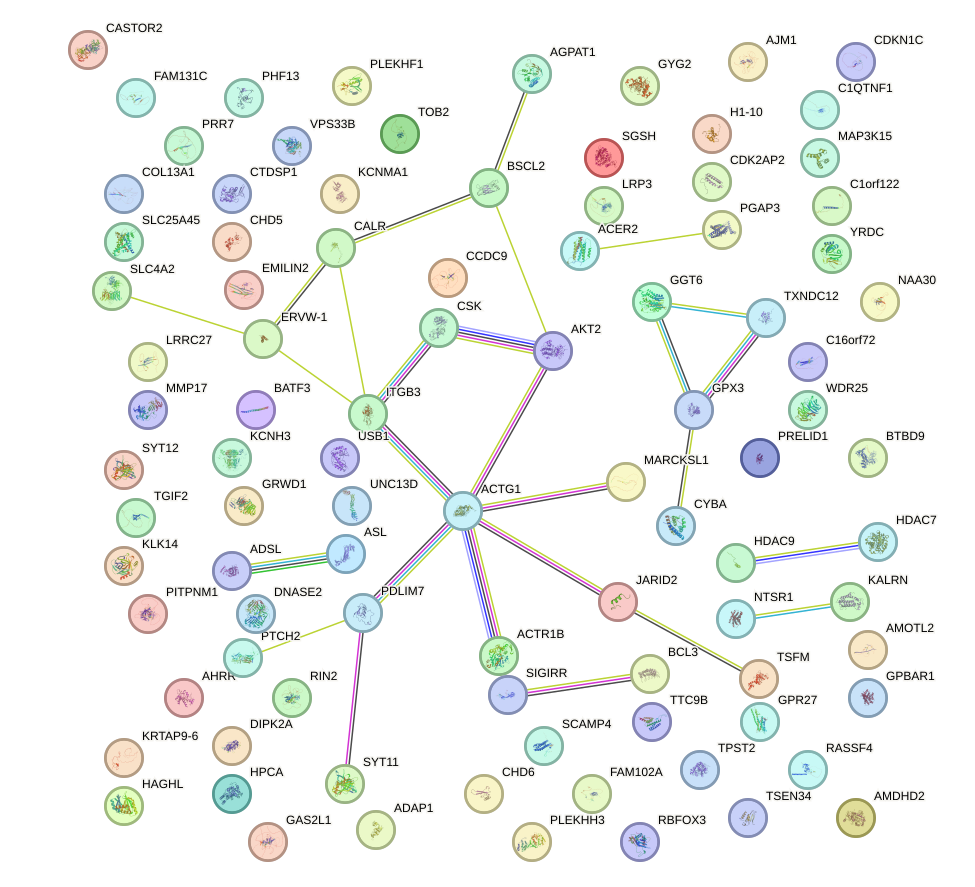
Network of associatons between targets according to the STRING database.
First level regulatory network of ZIC3_ZIC4
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 0.0 | 0.2 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.0 | 0.1 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.0 | 0.2 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.0 | 0.2 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.0 | 0.1 | GO:1904845 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.0 | 0.1 | GO:0097473 | cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.0 | 0.1 | GO:0006550 | isoleucine catabolic process(GO:0006550) |
| 0.0 | 0.1 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 0.1 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.0 | 0.1 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 0.0 | 0.1 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.1 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.1 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 0.0 | GO:2000646 | lipid transport involved in lipid storage(GO:0010877) regulation of Cdc42 protein signal transduction(GO:0032489) positive regulation of receptor catabolic process(GO:2000646) |
| 0.0 | 0.0 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.0 | 0.0 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.0 | 0.1 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.1 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.0 | GO:0036466 | synaptic vesicle recycling via endosome(GO:0036466) |
| 0.0 | 0.1 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.0 | 0.1 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.0 | 0.1 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.0 | 0.1 | GO:0042756 | drinking behavior(GO:0042756) |
| 0.0 | 0.0 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.0 | 0.0 | GO:1901536 | negative regulation of DNA demethylation(GO:1901536) |
| 0.0 | 0.1 | GO:0002396 | MHC protein complex assembly(GO:0002396) peptide antigen assembly with MHC protein complex(GO:0002501) |
| 0.0 | 0.0 | GO:0042662 | negative regulation of mesodermal cell fate specification(GO:0042662) cardiac cell fate determination(GO:0060913) |
| 0.0 | 0.0 | GO:1905224 | clathrin-coated pit assembly(GO:1905224) |
| 0.0 | 0.1 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.1 | GO:1902075 | cellular response to salt(GO:1902075) |
| 0.0 | 0.1 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0030936 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.0 | 0.1 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.1 | GO:0034753 | nuclear aryl hydrocarbon receptor complex(GO:0034753) |
| 0.0 | 0.1 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.1 | GO:0036338 | viral envelope(GO:0019031) viral membrane(GO:0036338) |
| 0.0 | 0.0 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.1 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.0 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.0 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.0 | 0.2 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0016250 | N-sulfoglucosamine sulfohydrolase activity(GO:0016250) hydrolase activity, acting on acid sulfur-nitrogen bonds(GO:0016826) |
| 0.1 | 0.2 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
| 0.0 | 0.1 | GO:0008466 | glycogenin glucosyltransferase activity(GO:0008466) |
| 0.0 | 0.1 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 0.2 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.0 | 0.0 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.0 | GO:0016781 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.0 | 0.1 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.1 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.1 | GO:0052655 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.0 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.2 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.1 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.1 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.1 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |