Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
Results for ZIC2_GLI1
Z-value: 0.77
Transcription factors associated with ZIC2_GLI1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
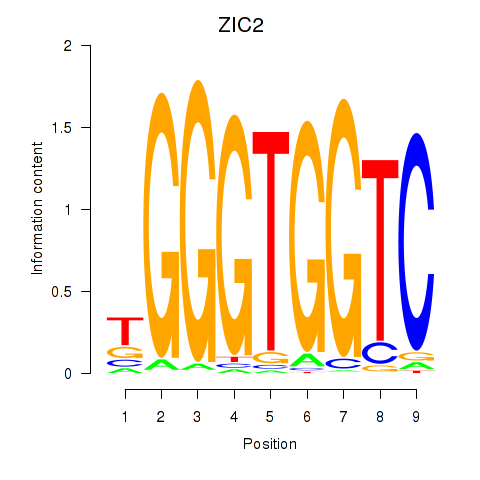
ZIC2
|
ENSG00000043355.6 | Zic family member 2 |
|
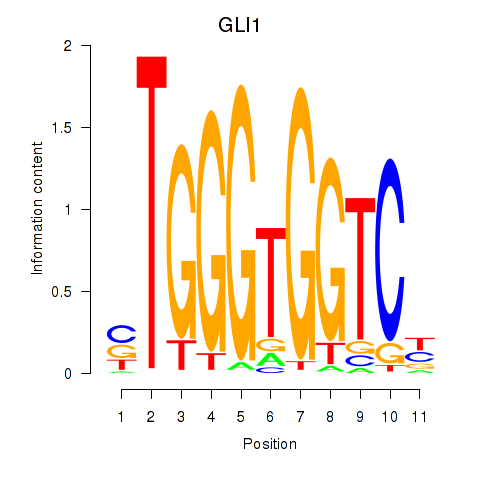
GLI1
|
ENSG00000111087.5 | GLI family zinc finger 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GLI1 | hg19_v2_chr12_+_57857475_57857475 | -0.40 | 6.0e-01 | Click! |
| ZIC2 | hg19_v2_chr13_+_100634004_100634026 | -0.38 | 6.2e-01 | Click! |
Activity profile of ZIC2_GLI1 motif
Sorted Z-values of ZIC2_GLI1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_26027480 | 1.02 |
ENST00000377364.3
|
HIST1H4B
|
histone cluster 1, H4b |
| chr16_-_21452040 | 0.45 |
ENST00000521589.1
|
NPIPB3
|
nuclear pore complex interacting protein family, member B3 |
| chr20_+_36149602 | 0.40 |
ENST00000062104.2
ENST00000346199.2 |
NNAT
|
neuronatin |
| chr1_+_209941942 | 0.29 |
ENST00000487271.1
ENST00000477431.1 |
TRAF3IP3
|
TRAF3 interacting protein 3 |
| chr16_+_86612112 | 0.25 |
ENST00000320241.3
|
FOXL1
|
forkhead box L1 |
| chrX_+_47444613 | 0.25 |
ENST00000445623.1
|
TIMP1
|
TIMP metallopeptidase inhibitor 1 |
| chr15_-_83224682 | 0.22 |
ENST00000562833.1
|
RP11-152F13.10
|
RP11-152F13.10 |
| chr17_+_73089382 | 0.19 |
ENST00000538213.2
ENST00000584118.1 |
SLC16A5
|
solute carrier family 16 (monocarboxylate transporter), member 5 |
| chr20_-_62493217 | 0.18 |
ENST00000601296.1
|
C20ORF135
|
C20ORF135 |
| chr1_-_45308616 | 0.18 |
ENST00000447098.2
ENST00000372192.3 |
PTCH2
|
patched 2 |
| chr20_+_57430162 | 0.18 |
ENST00000450130.1
ENST00000349036.3 ENST00000423897.1 |
GNAS
|
GNAS complex locus |
| chr3_-_10028366 | 0.17 |
ENST00000429759.1
|
EMC3
|
ER membrane protein complex subunit 3 |
| chr15_-_78526855 | 0.17 |
ENST00000541759.1
ENST00000558130.1 |
ACSBG1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr1_-_63782888 | 0.17 |
ENST00000436475.2
|
LINC00466
|
long intergenic non-protein coding RNA 466 |
| chr20_+_32319463 | 0.16 |
ENST00000342427.2
ENST00000375200.1 |
ZNF341
|
zinc finger protein 341 |
| chr12_+_65996599 | 0.16 |
ENST00000539116.1
ENST00000541391.1 |
RP11-221N13.3
|
RP11-221N13.3 |
| chr5_+_52776449 | 0.15 |
ENST00000396947.3
|
FST
|
follistatin |
| chr16_-_1031259 | 0.14 |
ENST00000563837.1
ENST00000563863.1 ENST00000565069.1 ENST00000570014.1 |
RP11-161M6.2
LMF1
|
RP11-161M6.2 lipase maturation factor 1 |
| chr22_-_50970506 | 0.14 |
ENST00000428989.2
ENST00000403326.1 |
ODF3B
|
outer dense fiber of sperm tails 3B |
| chr2_+_97481974 | 0.14 |
ENST00000377060.3
ENST00000305510.3 |
CNNM3
|
cyclin M3 |
| chr6_-_29600559 | 0.13 |
ENST00000476670.1
|
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr12_-_6798523 | 0.13 |
ENST00000319770.3
|
ZNF384
|
zinc finger protein 384 |
| chr11_-_64527425 | 0.13 |
ENST00000377432.3
|
PYGM
|
phosphorylase, glycogen, muscle |
| chr14_+_24583836 | 0.13 |
ENST00000559115.1
ENST00000558215.1 ENST00000557810.1 ENST00000561375.1 ENST00000446197.3 ENST00000559796.1 ENST00000560713.1 ENST00000560901.1 ENST00000559382.1 |
DCAF11
|
DDB1 and CUL4 associated factor 11 |
| chr14_+_64971438 | 0.12 |
ENST00000555321.1
|
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr3_-_16554403 | 0.12 |
ENST00000449415.1
ENST00000441460.1 |
RFTN1
|
raftlin, lipid raft linker 1 |
| chr1_-_205782304 | 0.11 |
ENST00000367137.3
|
SLC41A1
|
solute carrier family 41 (magnesium transporter), member 1 |
| chr6_+_43739697 | 0.11 |
ENST00000230480.6
|
VEGFA
|
vascular endothelial growth factor A |
| chr21_+_34804479 | 0.11 |
ENST00000421802.1
|
IFNGR2
|
interferon gamma receptor 2 (interferon gamma transducer 1) |
| chr10_+_99344071 | 0.10 |
ENST00000370647.4
ENST00000370646.4 |
HOGA1
|
4-hydroxy-2-oxoglutarate aldolase 1 |
| chr20_+_62327996 | 0.10 |
ENST00000369996.1
|
TNFRSF6B
|
tumor necrosis factor receptor superfamily, member 6b, decoy |
| chr6_+_31620191 | 0.10 |
ENST00000375918.2
ENST00000375920.4 |
APOM
|
apolipoprotein M |
| chr17_+_73997419 | 0.10 |
ENST00000425876.2
|
CDK3
|
cyclin-dependent kinase 3 |
| chr20_+_35202909 | 0.09 |
ENST00000558530.1
ENST00000558028.1 ENST00000560025.1 |
TGIF2-C20orf24
TGIF2
|
TGIF2-C20orf24 readthrough TGFB-induced factor homeobox 2 |
| chr7_+_69064300 | 0.09 |
ENST00000342771.4
|
AUTS2
|
autism susceptibility candidate 2 |
| chr14_-_61124977 | 0.09 |
ENST00000554986.1
|
SIX1
|
SIX homeobox 1 |
| chr19_-_44285401 | 0.09 |
ENST00000262888.3
|
KCNN4
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
| chr8_-_22014339 | 0.09 |
ENST00000306317.2
|
LGI3
|
leucine-rich repeat LGI family, member 3 |
| chr11_+_117049910 | 0.09 |
ENST00000431081.2
ENST00000524842.1 |
SIDT2
|
SID1 transmembrane family, member 2 |
| chr8_+_31497271 | 0.09 |
ENST00000520407.1
|
NRG1
|
neuregulin 1 |
| chr12_+_104337515 | 0.09 |
ENST00000550595.1
|
HSP90B1
|
heat shock protein 90kDa beta (Grp94), member 1 |
| chr12_+_31477250 | 0.09 |
ENST00000313737.4
|
AC024940.1
|
AC024940.1 |
| chr16_+_57844549 | 0.09 |
ENST00000564282.1
|
CTD-2600O9.1
|
uncharacterized protein LOC388282 |
| chr1_-_182573514 | 0.09 |
ENST00000367558.5
|
RGS16
|
regulator of G-protein signaling 16 |
| chr5_+_52776228 | 0.08 |
ENST00000256759.3
|
FST
|
follistatin |
| chrX_+_12809463 | 0.08 |
ENST00000380663.3
ENST00000380668.5 ENST00000398491.2 ENST00000489404.1 |
PRPS2
|
phosphoribosyl pyrophosphate synthetase 2 |
| chr5_+_175792459 | 0.08 |
ENST00000310389.5
|
ARL10
|
ADP-ribosylation factor-like 10 |
| chr22_-_46373004 | 0.08 |
ENST00000339464.4
|
WNT7B
|
wingless-type MMTV integration site family, member 7B |
| chr22_-_50970566 | 0.08 |
ENST00000405135.1
ENST00000401779.1 |
ODF3B
|
outer dense fiber of sperm tails 3B |
| chrX_+_152240819 | 0.08 |
ENST00000421798.3
ENST00000535416.1 |
PNMA6C
PNMA6A
|
paraneoplastic Ma antigen family member 6C paraneoplastic Ma antigen family member 6A |
| chr22_-_50970919 | 0.08 |
ENST00000329363.4
ENST00000437588.1 |
ODF3B
|
outer dense fiber of sperm tails 3B |
| chr20_+_57427765 | 0.08 |
ENST00000371100.4
|
GNAS
|
GNAS complex locus |
| chr12_-_49333446 | 0.08 |
ENST00000537495.1
|
AC073610.5
|
Uncharacterized protein |
| chr12_-_6798616 | 0.07 |
ENST00000355772.4
ENST00000417772.3 ENST00000396801.3 ENST00000396799.2 |
ZNF384
|
zinc finger protein 384 |
| chr22_+_38035459 | 0.07 |
ENST00000357436.4
|
SH3BP1
|
SH3-domain binding protein 1 |
| chr5_+_71403280 | 0.07 |
ENST00000511641.2
|
MAP1B
|
microtubule-associated protein 1B |
| chr1_+_183155373 | 0.07 |
ENST00000493293.1
ENST00000264144.4 |
LAMC2
|
laminin, gamma 2 |
| chr1_+_44679370 | 0.07 |
ENST00000372290.4
|
DMAP1
|
DNA methyltransferase 1 associated protein 1 |
| chr17_-_15496722 | 0.07 |
ENST00000472534.1
|
CDRT1
|
CMT1A duplicated region transcript 1 |
| chr4_-_176733897 | 0.07 |
ENST00000393658.2
|
GPM6A
|
glycoprotein M6A |
| chr16_+_2521500 | 0.07 |
ENST00000293973.1
|
NTN3
|
netrin 3 |
| chr3_+_113616317 | 0.07 |
ENST00000440446.2
ENST00000488680.1 |
GRAMD1C
|
GRAM domain containing 1C |
| chr22_-_19165917 | 0.07 |
ENST00000451283.1
|
SLC25A1
|
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1 |
| chr1_+_24120143 | 0.07 |
ENST00000374501.1
|
LYPLA2
|
lysophospholipase II |
| chr12_-_129308041 | 0.07 |
ENST00000376740.4
|
SLC15A4
|
solute carrier family 15 (oligopeptide transporter), member 4 |
| chr11_-_64901900 | 0.07 |
ENST00000526060.1
ENST00000307289.6 ENST00000528487.1 |
SYVN1
|
synovial apoptosis inhibitor 1, synoviolin |
| chr19_-_40919271 | 0.07 |
ENST00000291825.7
ENST00000324001.7 |
PRX
|
periaxin |
| chr1_-_40105617 | 0.07 |
ENST00000372852.3
|
HEYL
|
hes-related family bHLH transcription factor with YRPW motif-like |
| chr1_-_171621815 | 0.07 |
ENST00000037502.6
|
MYOC
|
myocilin, trabecular meshwork inducible glucocorticoid response |
| chr20_-_50722183 | 0.07 |
ENST00000371523.4
|
ZFP64
|
ZFP64 zinc finger protein |
| chr10_-_97050777 | 0.06 |
ENST00000329399.6
|
PDLIM1
|
PDZ and LIM domain 1 |
| chr15_-_43882140 | 0.06 |
ENST00000429176.1
|
PPIP5K1
|
diphosphoinositol pentakisphosphate kinase 1 |
| chr2_-_28113965 | 0.06 |
ENST00000302188.3
|
RBKS
|
ribokinase |
| chr19_+_1383890 | 0.06 |
ENST00000539480.1
ENST00000313408.7 ENST00000414651.2 |
NDUFS7
|
NADH dehydrogenase (ubiquinone) Fe-S protein 7, 20kDa (NADH-coenzyme Q reductase) |
| chr17_+_38497640 | 0.06 |
ENST00000394086.3
|
RARA
|
retinoic acid receptor, alpha |
| chr16_-_68002456 | 0.06 |
ENST00000576616.1
ENST00000572037.1 ENST00000338335.3 ENST00000422611.2 ENST00000316341.3 |
SLC12A4
|
solute carrier family 12 (potassium/chloride transporter), member 4 |
| chr14_+_24584372 | 0.06 |
ENST00000559396.1
ENST00000558638.1 ENST00000561041.1 ENST00000559288.1 ENST00000558408.1 |
DCAF11
|
DDB1 and CUL4 associated factor 11 |
| chr16_+_29818857 | 0.06 |
ENST00000567444.1
|
MAZ
|
MYC-associated zinc finger protein (purine-binding transcription factor) |
| chr11_-_64901978 | 0.06 |
ENST00000294256.8
ENST00000377190.3 |
SYVN1
|
synovial apoptosis inhibitor 1, synoviolin |
| chr20_+_44637526 | 0.06 |
ENST00000372330.3
|
MMP9
|
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) |
| chr11_-_57103327 | 0.06 |
ENST00000529002.1
ENST00000278412.2 |
SSRP1
|
structure specific recognition protein 1 |
| chr14_-_64971288 | 0.06 |
ENST00000394715.1
|
ZBTB25
|
zinc finger and BTB domain containing 25 |
| chr4_-_102268628 | 0.06 |
ENST00000323055.6
ENST00000512215.1 ENST00000394854.3 |
PPP3CA
|
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr3_+_47844615 | 0.06 |
ENST00000348968.4
|
DHX30
|
DEAH (Asp-Glu-Ala-His) box helicase 30 |
| chr2_-_31360887 | 0.06 |
ENST00000420311.2
ENST00000356174.3 ENST00000324589.5 |
GALNT14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chr17_+_7211280 | 0.06 |
ENST00000419711.2
ENST00000571955.1 ENST00000573714.1 |
EIF5A
|
eukaryotic translation initiation factor 5A |
| chr3_+_88108381 | 0.06 |
ENST00000473136.1
|
RP11-159G9.5
|
Uncharacterized protein |
| chr9_-_35103105 | 0.06 |
ENST00000452248.2
ENST00000356493.5 |
STOML2
|
stomatin (EPB72)-like 2 |
| chr14_+_92980111 | 0.06 |
ENST00000216487.7
ENST00000557762.1 |
RIN3
|
Ras and Rab interactor 3 |
| chr16_-_85146040 | 0.06 |
ENST00000539556.1
|
FAM92B
|
family with sequence similarity 92, member B |
| chr15_-_65281775 | 0.06 |
ENST00000433215.2
ENST00000558415.1 ENST00000557795.1 |
SPG21
|
spastic paraplegia 21 (autosomal recessive, Mast syndrome) |
| chr17_+_76422409 | 0.06 |
ENST00000600087.1
|
AC061992.1
|
Uncharacterized protein |
| chr9_-_130639997 | 0.06 |
ENST00000373176.1
|
AK1
|
adenylate kinase 1 |
| chr3_-_88108192 | 0.05 |
ENST00000309534.6
|
CGGBP1
|
CGG triplet repeat binding protein 1 |
| chr6_-_32821599 | 0.05 |
ENST00000354258.4
|
TAP1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr5_-_168727786 | 0.05 |
ENST00000332966.8
|
SLIT3
|
slit homolog 3 (Drosophila) |
| chr10_-_98347063 | 0.05 |
ENST00000443638.1
|
TM9SF3
|
transmembrane 9 superfamily member 3 |
| chr1_-_211752073 | 0.05 |
ENST00000367001.4
|
SLC30A1
|
solute carrier family 30 (zinc transporter), member 1 |
| chr11_-_111783919 | 0.05 |
ENST00000531198.1
ENST00000533879.1 |
CRYAB
|
crystallin, alpha B |
| chr21_-_34100341 | 0.05 |
ENST00000382499.2
ENST00000433931.2 |
SYNJ1
|
synaptojanin 1 |
| chr9_-_98079965 | 0.05 |
ENST00000289081.3
|
FANCC
|
Fanconi anemia, complementation group C |
| chr10_-_95360983 | 0.05 |
ENST00000371464.3
|
RBP4
|
retinol binding protein 4, plasma |
| chr5_-_141704566 | 0.05 |
ENST00000344120.4
ENST00000434127.2 |
SPRY4
|
sprouty homolog 4 (Drosophila) |
| chr4_+_72052964 | 0.05 |
ENST00000264485.5
ENST00000425175.1 |
SLC4A4
|
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
| chr12_-_6798410 | 0.05 |
ENST00000361959.3
ENST00000436774.2 ENST00000544482.1 |
ZNF384
|
zinc finger protein 384 |
| chr2_-_51259641 | 0.05 |
ENST00000406316.2
ENST00000405581.1 |
NRXN1
|
neurexin 1 |
| chr17_-_7137582 | 0.05 |
ENST00000575756.1
ENST00000575458.1 |
DVL2
|
dishevelled segment polarity protein 2 |
| chr3_+_47844399 | 0.05 |
ENST00000446256.2
ENST00000445061.1 |
DHX30
|
DEAH (Asp-Glu-Ala-His) box helicase 30 |
| chrX_+_51928002 | 0.05 |
ENST00000375626.3
|
MAGED4
|
melanoma antigen family D, 4 |
| chrX_-_107334790 | 0.05 |
ENST00000217958.3
|
PSMD10
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 10 |
| chr11_+_33278811 | 0.05 |
ENST00000303296.4
ENST00000379016.3 |
HIPK3
|
homeodomain interacting protein kinase 3 |
| chr11_-_75379612 | 0.05 |
ENST00000526740.1
|
MAP6
|
microtubule-associated protein 6 |
| chr21_-_34100244 | 0.05 |
ENST00000382491.3
ENST00000357345.3 ENST00000429236.1 |
SYNJ1
|
synaptojanin 1 |
| chr19_+_36208877 | 0.05 |
ENST00000420124.1
ENST00000222270.7 ENST00000341701.1 |
KMT2B
|
Histone-lysine N-methyltransferase 2B |
| chr9_-_8857776 | 0.05 |
ENST00000481079.1
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D |
| chr13_-_30880979 | 0.05 |
ENST00000414289.1
|
KATNAL1
|
katanin p60 subunit A-like 1 |
| chr7_-_105516923 | 0.05 |
ENST00000478915.1
|
ATXN7L1
|
ataxin 7-like 1 |
| chr3_+_137717571 | 0.05 |
ENST00000343735.4
|
CLDN18
|
claudin 18 |
| chr15_-_65282274 | 0.05 |
ENST00000204566.2
|
SPG21
|
spastic paraplegia 21 (autosomal recessive, Mast syndrome) |
| chr22_+_38035623 | 0.05 |
ENST00000336738.5
ENST00000442465.2 |
SH3BP1
|
SH3-domain binding protein 1 |
| chr9_-_116159635 | 0.05 |
ENST00000452726.1
|
ALAD
|
aminolevulinate dehydratase |
| chrX_+_51636629 | 0.05 |
ENST00000375722.1
ENST00000326587.7 ENST00000375695.2 |
MAGED1
|
melanoma antigen family D, 1 |
| chr17_-_37558776 | 0.04 |
ENST00000577399.1
|
FBXL20
|
F-box and leucine-rich repeat protein 20 |
| chr22_-_28197486 | 0.04 |
ENST00000302326.4
|
MN1
|
meningioma (disrupted in balanced translocation) 1 |
| chr1_+_44679159 | 0.04 |
ENST00000315913.5
ENST00000372289.2 |
DMAP1
|
DNA methyltransferase 1 associated protein 1 |
| chr7_-_106301405 | 0.04 |
ENST00000523505.1
|
CCDC71L
|
coiled-coil domain containing 71-like |
| chr8_+_22225041 | 0.04 |
ENST00000289952.5
ENST00000524285.1 |
SLC39A14
|
solute carrier family 39 (zinc transporter), member 14 |
| chr2_-_133427767 | 0.04 |
ENST00000397463.2
|
LYPD1
|
LY6/PLAUR domain containing 1 |
| chr1_-_12677714 | 0.04 |
ENST00000376223.2
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr19_-_39330818 | 0.04 |
ENST00000594769.1
ENST00000602021.1 |
AC104534.3
|
Delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase, mitochondrial |
| chr9_-_98279241 | 0.04 |
ENST00000437951.1
ENST00000375274.2 ENST00000430669.2 ENST00000468211.2 |
PTCH1
|
patched 1 |
| chr17_-_26989136 | 0.04 |
ENST00000247020.4
|
SDF2
|
stromal cell-derived factor 2 |
| chr6_-_31620455 | 0.04 |
ENST00000437771.1
ENST00000404765.2 ENST00000375964.6 ENST00000211379.5 |
BAG6
|
BCL2-associated athanogene 6 |
| chrX_-_140271249 | 0.04 |
ENST00000370526.2
|
LDOC1
|
leucine zipper, down-regulated in cancer 1 |
| chr2_+_106361333 | 0.04 |
ENST00000233154.4
ENST00000451463.2 |
NCK2
|
NCK adaptor protein 2 |
| chr16_+_28834303 | 0.04 |
ENST00000340394.8
ENST00000325215.6 ENST00000395547.2 ENST00000336783.4 ENST00000382686.4 ENST00000564304.1 |
ATXN2L
|
ataxin 2-like |
| chr15_-_26108355 | 0.04 |
ENST00000356865.6
|
ATP10A
|
ATPase, class V, type 10A |
| chr12_-_25801478 | 0.04 |
ENST00000540106.1
ENST00000445693.1 ENST00000545543.1 ENST00000542224.1 |
IFLTD1
|
intermediate filament tail domain containing 1 |
| chr1_+_6673745 | 0.04 |
ENST00000377648.4
|
PHF13
|
PHD finger protein 13 |
| chr5_-_150460914 | 0.04 |
ENST00000389378.2
|
TNIP1
|
TNFAIP3 interacting protein 1 |
| chr8_+_30244580 | 0.04 |
ENST00000523115.1
ENST00000519647.1 |
RBPMS
|
RNA binding protein with multiple splicing |
| chr19_-_42758040 | 0.04 |
ENST00000593944.1
|
ERF
|
Ets2 repressor factor |
| chr9_+_109625378 | 0.04 |
ENST00000277225.5
ENST00000457913.1 ENST00000472574.1 |
ZNF462
|
zinc finger protein 462 |
| chr1_-_161014731 | 0.04 |
ENST00000368020.1
|
USF1
|
upstream transcription factor 1 |
| chr11_-_65321198 | 0.04 |
ENST00000530426.1
|
LTBP3
|
latent transforming growth factor beta binding protein 3 |
| chr6_+_20403997 | 0.04 |
ENST00000535432.1
|
E2F3
|
E2F transcription factor 3 |
| chr12_+_119419294 | 0.04 |
ENST00000267260.4
|
SRRM4
|
serine/arginine repetitive matrix 4 |
| chr19_+_50354462 | 0.04 |
ENST00000601675.1
|
PTOV1
|
prostate tumor overexpressed 1 |
| chr10_+_18948311 | 0.04 |
ENST00000377275.3
|
ARL5B
|
ADP-ribosylation factor-like 5B |
| chr6_+_125540951 | 0.04 |
ENST00000524679.1
|
TPD52L1
|
tumor protein D52-like 1 |
| chr17_-_15244894 | 0.04 |
ENST00000338696.2
ENST00000543896.1 ENST00000539245.1 ENST00000539316.1 ENST00000395930.1 |
TEKT3
|
tektin 3 |
| chr3_+_67048721 | 0.04 |
ENST00000295568.4
ENST00000484414.1 ENST00000460576.1 ENST00000417314.2 |
KBTBD8
|
kelch repeat and BTB (POZ) domain containing 8 |
| chr11_-_66104237 | 0.04 |
ENST00000530056.1
|
RIN1
|
Ras and Rab interactor 1 |
| chr6_+_160183492 | 0.03 |
ENST00000541436.1
|
ACAT2
|
acetyl-CoA acetyltransferase 2 |
| chr2_-_85829496 | 0.03 |
ENST00000409668.1
|
TMEM150A
|
transmembrane protein 150A |
| chr2_+_28113583 | 0.03 |
ENST00000344773.2
ENST00000379624.1 ENST00000342045.2 ENST00000379632.2 ENST00000361704.2 |
BRE
|
brain and reproductive organ-expressed (TNFRSF1A modulator) |
| chr1_+_209941827 | 0.03 |
ENST00000367023.1
|
TRAF3IP3
|
TRAF3 interacting protein 3 |
| chr11_+_45944190 | 0.03 |
ENST00000401752.1
ENST00000389968.3 ENST00000325468.5 ENST00000536139.1 |
GYLTL1B
|
glycosyltransferase-like 1B |
| chr21_-_28217721 | 0.03 |
ENST00000284984.3
|
ADAMTS1
|
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
| chr20_+_2082494 | 0.03 |
ENST00000246032.3
|
STK35
|
serine/threonine kinase 35 |
| chr10_-_77161004 | 0.03 |
ENST00000418818.2
|
RP11-399K21.11
|
RP11-399K21.11 |
| chr2_-_46769694 | 0.03 |
ENST00000522587.1
|
ATP6V1E2
|
ATPase, H+ transporting, lysosomal 31kDa, V1 subunit E2 |
| chr5_-_168727713 | 0.03 |
ENST00000404867.3
|
SLIT3
|
slit homolog 3 (Drosophila) |
| chr15_+_40861487 | 0.03 |
ENST00000315616.7
ENST00000559271.1 |
RPUSD2
|
RNA pseudouridylate synthase domain containing 2 |
| chr1_-_43833628 | 0.03 |
ENST00000413844.2
ENST00000372458.3 |
ELOVL1
|
ELOVL fatty acid elongase 1 |
| chr1_+_36621529 | 0.03 |
ENST00000316156.4
|
MAP7D1
|
MAP7 domain containing 1 |
| chr3_-_49449521 | 0.03 |
ENST00000431929.1
ENST00000418115.1 |
RHOA
|
ras homolog family member A |
| chr3_+_110790590 | 0.03 |
ENST00000485303.1
|
PVRL3
|
poliovirus receptor-related 3 |
| chr8_-_17555164 | 0.03 |
ENST00000297488.6
|
MTUS1
|
microtubule associated tumor suppressor 1 |
| chr11_-_111782696 | 0.03 |
ENST00000227251.3
ENST00000526180.1 |
CRYAB
|
crystallin, alpha B |
| chr17_+_73997796 | 0.03 |
ENST00000586261.1
|
CDK3
|
cyclin-dependent kinase 3 |
| chr14_+_77228532 | 0.03 |
ENST00000167106.4
ENST00000554237.1 |
VASH1
|
vasohibin 1 |
| chr5_+_161275320 | 0.03 |
ENST00000437025.2
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr3_+_137728842 | 0.03 |
ENST00000183605.5
|
CLDN18
|
claudin 18 |
| chr22_-_30783356 | 0.03 |
ENST00000382363.3
|
RNF215
|
ring finger protein 215 |
| chr15_-_78526942 | 0.03 |
ENST00000258873.4
|
ACSBG1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr15_-_65282232 | 0.03 |
ENST00000416889.2
|
SPG21
|
spastic paraplegia 21 (autosomal recessive, Mast syndrome) |
| chr1_+_108918421 | 0.03 |
ENST00000444143.2
ENST00000294652.8 |
NBPF6
|
neuroblastoma breakpoint family, member 6 |
| chr6_-_29600832 | 0.03 |
ENST00000377016.4
ENST00000376977.3 ENST00000377034.4 |
GABBR1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr3_-_136471204 | 0.03 |
ENST00000480733.1
ENST00000383202.2 ENST00000236698.5 ENST00000434713.2 |
STAG1
|
stromal antigen 1 |
| chr17_+_46133307 | 0.03 |
ENST00000580037.1
|
NFE2L1
|
nuclear factor, erythroid 2-like 1 |
| chr11_-_12030629 | 0.03 |
ENST00000396505.2
|
DKK3
|
dickkopf WNT signaling pathway inhibitor 3 |
| chr11_+_64001962 | 0.03 |
ENST00000309422.2
|
VEGFB
|
vascular endothelial growth factor B |
| chr12_+_100661156 | 0.03 |
ENST00000360820.2
|
SCYL2
|
SCY1-like 2 (S. cerevisiae) |
| chr17_-_7137857 | 0.03 |
ENST00000005340.5
|
DVL2
|
dishevelled segment polarity protein 2 |
| chr17_-_76778339 | 0.03 |
ENST00000591455.1
ENST00000446868.3 ENST00000361101.4 ENST00000589296.1 |
CYTH1
|
cytohesin 1 |
| chr22_-_19166343 | 0.03 |
ENST00000215882.5
|
SLC25A1
|
solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1 |
| chr17_+_4843679 | 0.02 |
ENST00000576229.1
|
RNF167
|
ring finger protein 167 |
| chr19_-_14629224 | 0.02 |
ENST00000254322.2
|
DNAJB1
|
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr19_+_4969116 | 0.02 |
ENST00000588337.1
ENST00000159111.4 ENST00000381759.4 |
KDM4B
|
lysine (K)-specific demethylase 4B |
| chr19_-_55881741 | 0.02 |
ENST00000264563.2
ENST00000590625.1 ENST00000585513.1 |
IL11
|
interleukin 11 |
| chr12_+_57854274 | 0.02 |
ENST00000528432.1
|
GLI1
|
GLI family zinc finger 1 |
| chr9_-_136242909 | 0.02 |
ENST00000371991.3
ENST00000545297.1 |
SURF4
|
surfeit 4 |
| chr6_-_31620403 | 0.02 |
ENST00000451898.1
ENST00000439687.2 ENST00000362049.6 ENST00000424480.1 |
BAG6
|
BCL2-associated athanogene 6 |
| chr6_-_31620149 | 0.02 |
ENST00000435080.1
ENST00000375976.4 ENST00000441054.1 |
BAG6
|
BCL2-associated athanogene 6 |
| chr7_-_150777874 | 0.02 |
ENST00000540185.1
|
FASTK
|
Fas-activated serine/threonine kinase |
| chr15_-_101142401 | 0.02 |
ENST00000314742.8
|
LINS
|
lines homolog (Drosophila) |
| chr16_+_56899114 | 0.02 |
ENST00000566786.1
ENST00000438926.2 ENST00000563236.1 ENST00000262502.5 |
SLC12A3
|
solute carrier family 12 (sodium/chloride transporter), member 3 |
| chr1_-_115053781 | 0.02 |
ENST00000358465.2
ENST00000369543.2 |
TRIM33
|
tripartite motif containing 33 |
| chr22_-_30783075 | 0.02 |
ENST00000215798.6
|
RNF215
|
ring finger protein 215 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZIC2_GLI1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1903570 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.0 | 0.3 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.1 | GO:2000174 | regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 0.0 | 0.1 | GO:0034444 | regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) |
| 0.0 | 0.1 | GO:0035674 | tricarboxylic acid transmembrane transport(GO:0035674) |
| 0.0 | 0.4 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.1 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.0 | 0.1 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 | 0.3 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 0.1 | GO:1904978 | regulation of endosome organization(GO:1904978) |
| 0.0 | 0.1 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.0 | 0.1 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.0 | 0.2 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.1 | GO:2000824 | negative regulation of androgen receptor activity(GO:2000824) |
| 0.0 | 0.1 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.1 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.0 | 0.1 | GO:0061364 | apoptotic process involved in luteolysis(GO:0061364) |
| 0.0 | 0.2 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.1 | GO:0060535 | trachea cartilage morphogenesis(GO:0060535) inner medullary collecting duct development(GO:0072061) |
| 0.0 | 0.1 | GO:1901860 | positive regulation of mitochondrial DNA metabolic process(GO:1901860) |
| 0.0 | 0.1 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 0.1 | GO:0019470 | 4-hydroxyproline catabolic process(GO:0019470) |
| 0.0 | 0.1 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.2 | GO:0009957 | epidermal cell fate specification(GO:0009957) |
| 0.0 | 0.2 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.0 | 0.1 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.0 | 0.1 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.0 | 0.1 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.0 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.0 | 0.0 | GO:0071284 | cellular response to lead ion(GO:0071284) |
| 0.0 | 0.1 | GO:0033578 | protein glycosylation in Golgi(GO:0033578) |
| 0.0 | 0.1 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.0 | 0.1 | GO:0046541 | saliva secretion(GO:0046541) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.0 | 0.1 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.1 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.2 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.3 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.0 | 0.1 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.0 | GO:1904854 | proteasome core complex binding(GO:1904854) |
| 0.0 | 0.1 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.1 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.0 | 0.0 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.2 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.1 | GO:0015333 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |