Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
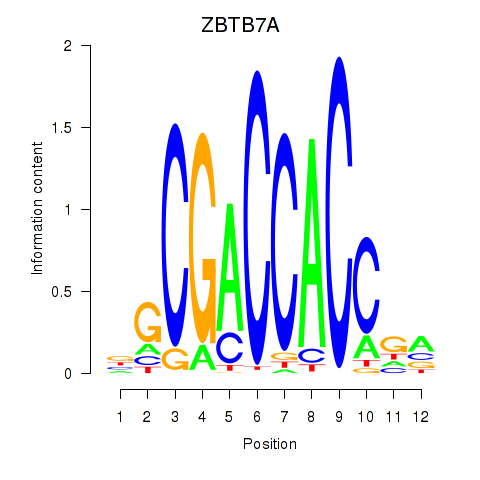
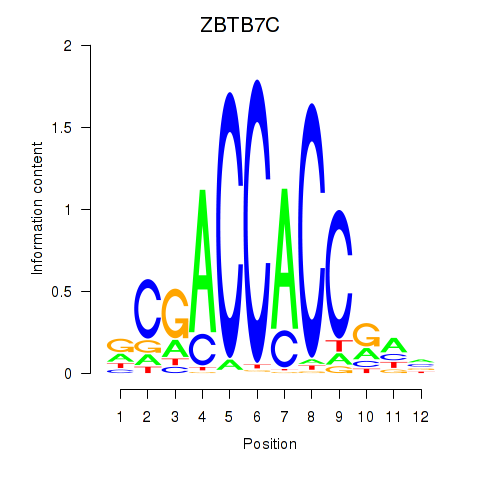
Results for ZBTB7A_ZBTB7C
Z-value: 0.60
Transcription factors associated with ZBTB7A_ZBTB7C
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB7A
|
ENSG00000178951.4 | zinc finger and BTB domain containing 7A |
|
ZBTB7C
|
ENSG00000184828.5 | zinc finger and BTB domain containing 7C |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZBTB7C | hg19_v2_chr18_-_45935663_45935793 | 0.93 | 6.6e-02 | Click! |
| ZBTB7A | hg19_v2_chr19_-_4066890_4066943 | -0.76 | 2.4e-01 | Click! |
Activity profile of ZBTB7A_ZBTB7C motif
Sorted Z-values of ZBTB7A_ZBTB7C motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_129872672 | 0.37 |
ENST00000531431.1
ENST00000527581.1 |
PRDM10
|
PR domain containing 10 |
| chr5_-_114515734 | 0.36 |
ENST00000514154.1
ENST00000282369.3 |
TRIM36
|
tripartite motif containing 36 |
| chr12_+_57854274 | 0.34 |
ENST00000528432.1
|
GLI1
|
GLI family zinc finger 1 |
| chr3_-_71803917 | 0.29 |
ENST00000421769.2
|
EIF4E3
|
eukaryotic translation initiation factor 4E family member 3 |
| chr5_-_138739739 | 0.27 |
ENST00000514983.1
ENST00000507779.2 ENST00000451821.2 ENST00000450845.2 ENST00000509959.1 ENST00000302091.5 |
SPATA24
|
spermatogenesis associated 24 |
| chr9_-_127905736 | 0.23 |
ENST00000336505.6
ENST00000373549.4 |
SCAI
|
suppressor of cancer cell invasion |
| chr3_-_71803474 | 0.19 |
ENST00000448225.1
ENST00000496214.2 |
EIF4E3
|
eukaryotic translation initiation factor 4E family member 3 |
| chr16_+_53164833 | 0.18 |
ENST00000564845.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr17_+_63096903 | 0.17 |
ENST00000582940.1
|
RP11-160O5.1
|
RP11-160O5.1 |
| chr14_-_102771462 | 0.17 |
ENST00000522874.1
|
MOK
|
MOK protein kinase |
| chr5_-_132112921 | 0.17 |
ENST00000378721.4
ENST00000378701.1 |
SEPT8
|
septin 8 |
| chr4_-_146101304 | 0.16 |
ENST00000447906.2
|
OTUD4
|
OTU domain containing 4 |
| chr17_-_5372068 | 0.16 |
ENST00000572490.1
|
DHX33
|
DEAH (Asp-Glu-Ala-His) box polypeptide 33 |
| chr16_+_2521500 | 0.16 |
ENST00000293973.1
|
NTN3
|
netrin 3 |
| chr17_-_8868991 | 0.16 |
ENST00000447110.1
|
PIK3R5
|
phosphoinositide-3-kinase, regulatory subunit 5 |
| chr7_-_94285472 | 0.15 |
ENST00000437425.2
ENST00000447873.1 ENST00000415788.2 |
SGCE
|
sarcoglycan, epsilon |
| chr7_-_94285402 | 0.15 |
ENST00000428696.2
ENST00000445866.2 |
SGCE
|
sarcoglycan, epsilon |
| chr14_-_102771516 | 0.14 |
ENST00000524214.1
ENST00000193029.6 ENST00000361847.2 |
MOK
|
MOK protein kinase |
| chr5_-_132112907 | 0.13 |
ENST00000458488.2
|
SEPT8
|
septin 8 |
| chr13_+_96204961 | 0.13 |
ENST00000299339.2
|
CLDN10
|
claudin 10 |
| chr5_-_132113063 | 0.13 |
ENST00000378719.2
|
SEPT8
|
septin 8 |
| chr17_-_46799872 | 0.13 |
ENST00000290294.3
|
PRAC1
|
prostate cancer susceptibility candidate 1 |
| chr11_-_12030681 | 0.13 |
ENST00000529338.1
|
DKK3
|
dickkopf WNT signaling pathway inhibitor 3 |
| chr4_-_56412713 | 0.13 |
ENST00000435527.2
|
CLOCK
|
clock circadian regulator |
| chr18_+_74207477 | 0.13 |
ENST00000532511.1
|
RP11-17M16.1
|
uncharacterized protein LOC400658 |
| chr4_-_47916543 | 0.13 |
ENST00000507489.1
|
NFXL1
|
nuclear transcription factor, X-box binding-like 1 |
| chr2_+_149402009 | 0.13 |
ENST00000457184.1
|
EPC2
|
enhancer of polycomb homolog 2 (Drosophila) |
| chr1_+_167905894 | 0.13 |
ENST00000367843.3
ENST00000432587.2 ENST00000312263.6 |
DCAF6
|
DDB1 and CUL4 associated factor 6 |
| chrX_-_48980098 | 0.13 |
ENST00000156109.5
|
GPKOW
|
G patch domain and KOW motifs |
| chr7_-_94285511 | 0.13 |
ENST00000265735.7
|
SGCE
|
sarcoglycan, epsilon |
| chr12_-_51611477 | 0.12 |
ENST00000389243.4
|
POU6F1
|
POU class 6 homeobox 1 |
| chr17_-_58469329 | 0.12 |
ENST00000393003.3
|
USP32
|
ubiquitin specific peptidase 32 |
| chr8_-_101963482 | 0.12 |
ENST00000419477.2
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr17_-_8534031 | 0.12 |
ENST00000411957.1
ENST00000396239.1 ENST00000379980.4 |
MYH10
|
myosin, heavy chain 10, non-muscle |
| chr11_+_71544246 | 0.12 |
ENST00000328698.1
|
DEFB108B
|
defensin, beta 108B |
| chr13_+_20532900 | 0.11 |
ENST00000382871.2
|
ZMYM2
|
zinc finger, MYM-type 2 |
| chr2_+_56411131 | 0.11 |
ENST00000407595.2
|
CCDC85A
|
coiled-coil domain containing 85A |
| chr5_+_76506706 | 0.11 |
ENST00000340978.3
ENST00000346042.3 ENST00000264917.5 ENST00000342343.4 ENST00000333194.4 |
PDE8B
|
phosphodiesterase 8B |
| chr5_+_102455968 | 0.11 |
ENST00000358359.3
|
PPIP5K2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr1_-_21616901 | 0.10 |
ENST00000436918.2
ENST00000374893.6 |
ECE1
|
endothelin converting enzyme 1 |
| chr9_-_123476612 | 0.10 |
ENST00000426959.1
|
MEGF9
|
multiple EGF-like-domains 9 |
| chr10_-_16859361 | 0.10 |
ENST00000377921.3
|
RSU1
|
Ras suppressor protein 1 |
| chr15_+_27112948 | 0.10 |
ENST00000555060.1
|
GABRA5
|
gamma-aminobutyric acid (GABA) A receptor, alpha 5 |
| chr3_+_179040767 | 0.10 |
ENST00000496856.1
ENST00000491818.1 |
ZNF639
|
zinc finger protein 639 |
| chr14_+_100150622 | 0.10 |
ENST00000261835.3
|
CYP46A1
|
cytochrome P450, family 46, subfamily A, polypeptide 1 |
| chr10_+_14880364 | 0.10 |
ENST00000441647.1
|
HSPA14
|
heat shock 70kDa protein 14 |
| chr7_+_77167376 | 0.09 |
ENST00000435495.2
|
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12 |
| chr11_-_17035943 | 0.09 |
ENST00000355661.3
ENST00000532079.1 ENST00000448080.2 ENST00000531066.1 |
PLEKHA7
|
pleckstrin homology domain containing, family A member 7 |
| chr13_-_30881134 | 0.09 |
ENST00000380617.3
ENST00000441394.1 |
KATNAL1
|
katanin p60 subunit A-like 1 |
| chr5_+_78407602 | 0.09 |
ENST00000274353.5
ENST00000524080.1 |
BHMT
|
betaine--homocysteine S-methyltransferase |
| chr17_-_56065484 | 0.09 |
ENST00000581208.1
|
VEZF1
|
vascular endothelial zinc finger 1 |
| chr1_-_156722195 | 0.09 |
ENST00000368206.5
|
HDGF
|
hepatoma-derived growth factor |
| chr16_+_84002234 | 0.09 |
ENST00000305202.4
|
NECAB2
|
N-terminal EF-hand calcium binding protein 2 |
| chr20_-_58508359 | 0.09 |
ENST00000446834.1
|
SYCP2
|
synaptonemal complex protein 2 |
| chr5_-_132113690 | 0.08 |
ENST00000414594.1
|
SEPT8
|
septin 8 |
| chr17_-_8534067 | 0.08 |
ENST00000360416.3
ENST00000269243.4 |
MYH10
|
myosin, heavy chain 10, non-muscle |
| chr2_+_27505260 | 0.08 |
ENST00000380075.2
ENST00000296098.4 |
TRIM54
|
tripartite motif containing 54 |
| chr17_-_39769005 | 0.08 |
ENST00000301653.4
ENST00000593067.1 |
KRT16
|
keratin 16 |
| chr19_-_50143452 | 0.08 |
ENST00000246792.3
|
RRAS
|
related RAS viral (r-ras) oncogene homolog |
| chr7_+_77167343 | 0.08 |
ENST00000433369.2
ENST00000415482.2 |
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12 |
| chr17_-_5372229 | 0.08 |
ENST00000433302.3
|
DHX33
|
DEAH (Asp-Glu-Ala-His) box polypeptide 33 |
| chr11_+_133938955 | 0.08 |
ENST00000534549.1
ENST00000441717.3 |
JAM3
|
junctional adhesion molecule 3 |
| chr12_+_57943781 | 0.08 |
ENST00000455537.2
ENST00000286452.5 |
KIF5A
|
kinesin family member 5A |
| chr12_-_56221330 | 0.08 |
ENST00000546837.1
|
RP11-762I7.5
|
Uncharacterized protein |
| chr9_+_74526384 | 0.08 |
ENST00000334731.2
ENST00000377031.3 |
C9orf85
|
chromosome 9 open reading frame 85 |
| chr1_+_182992545 | 0.08 |
ENST00000258341.4
|
LAMC1
|
laminin, gamma 1 (formerly LAMB2) |
| chr2_-_25016251 | 0.07 |
ENST00000328379.5
|
PTRHD1
|
peptidyl-tRNA hydrolase domain containing 1 |
| chr17_-_58469687 | 0.07 |
ENST00000590133.1
|
USP32
|
ubiquitin specific peptidase 32 |
| chrX_-_151143140 | 0.07 |
ENST00000393914.3
ENST00000370328.3 ENST00000370325.1 |
GABRE
|
gamma-aminobutyric acid (GABA) A receptor, epsilon |
| chr8_-_101963677 | 0.07 |
ENST00000395956.3
ENST00000395953.2 |
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr17_-_42298201 | 0.07 |
ENST00000527034.1
|
UBTF
|
upstream binding transcription factor, RNA polymerase I |
| chr4_-_7069760 | 0.07 |
ENST00000264954.4
|
GRPEL1
|
GrpE-like 1, mitochondrial (E. coli) |
| chr14_+_39644387 | 0.07 |
ENST00000553331.1
ENST00000216832.4 |
PNN
|
pinin, desmosome associated protein |
| chr4_-_119274121 | 0.07 |
ENST00000296498.3
|
PRSS12
|
protease, serine, 12 (neurotrypsin, motopsin) |
| chr8_-_101734170 | 0.07 |
ENST00000522387.1
ENST00000518196.1 |
PABPC1
|
poly(A) binding protein, cytoplasmic 1 |
| chr19_-_48673552 | 0.07 |
ENST00000536218.1
ENST00000596549.1 |
LIG1
|
ligase I, DNA, ATP-dependent |
| chrX_-_119603138 | 0.07 |
ENST00000200639.4
ENST00000371335.4 ENST00000538785.1 ENST00000434600.2 |
LAMP2
|
lysosomal-associated membrane protein 2 |
| chr2_+_48667983 | 0.06 |
ENST00000449090.2
|
PPP1R21
|
protein phosphatase 1, regulatory subunit 21 |
| chr12_-_120805872 | 0.06 |
ENST00000546985.1
|
MSI1
|
musashi RNA-binding protein 1 |
| chr2_+_136289030 | 0.06 |
ENST00000409478.1
ENST00000264160.4 ENST00000329971.3 ENST00000438014.1 |
R3HDM1
|
R3H domain containing 1 |
| chr2_+_46769798 | 0.06 |
ENST00000238738.4
|
RHOQ
|
ras homolog family member Q |
| chr5_-_145214848 | 0.06 |
ENST00000505416.1
ENST00000334744.4 ENST00000358004.2 ENST00000511435.1 |
PRELID2
|
PRELI domain containing 2 |
| chr2_+_37311588 | 0.06 |
ENST00000409774.1
ENST00000608836.1 |
GPATCH11
|
G patch domain containing 11 |
| chr1_-_202776392 | 0.06 |
ENST00000235790.4
|
KDM5B
|
lysine (K)-specific demethylase 5B |
| chr4_-_76598326 | 0.06 |
ENST00000503660.1
|
G3BP2
|
GTPase activating protein (SH3 domain) binding protein 2 |
| chr10_+_70091847 | 0.06 |
ENST00000441000.2
ENST00000354695.5 |
HNRNPH3
|
heterogeneous nuclear ribonucleoprotein H3 (2H9) |
| chr11_-_57102947 | 0.06 |
ENST00000526696.1
|
SSRP1
|
structure specific recognition protein 1 |
| chr11_-_65429891 | 0.06 |
ENST00000527874.1
|
RELA
|
v-rel avian reticuloendotheliosis viral oncogene homolog A |
| chr16_-_49315731 | 0.06 |
ENST00000219197.6
|
CBLN1
|
cerebellin 1 precursor |
| chr13_+_20532848 | 0.06 |
ENST00000382874.2
|
ZMYM2
|
zinc finger, MYM-type 2 |
| chr1_+_167906056 | 0.06 |
ENST00000367840.3
|
DCAF6
|
DDB1 and CUL4 associated factor 6 |
| chr17_-_56065540 | 0.06 |
ENST00000583932.1
|
VEZF1
|
vascular endothelial zinc finger 1 |
| chr19_-_10446449 | 0.06 |
ENST00000592439.1
|
ICAM3
|
intercellular adhesion molecule 3 |
| chr15_-_64126084 | 0.06 |
ENST00000560316.1
ENST00000443617.2 ENST00000560462.1 ENST00000558532.1 ENST00000561400.1 |
HERC1
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
| chr10_+_60144782 | 0.06 |
ENST00000487519.1
|
TFAM
|
transcription factor A, mitochondrial |
| chr12_-_57882498 | 0.05 |
ENST00000550288.1
|
ARHGAP9
|
Rho GTPase activating protein 9 |
| chr3_-_113415441 | 0.05 |
ENST00000491165.1
ENST00000316407.4 |
KIAA2018
|
KIAA2018 |
| chr17_-_5372271 | 0.05 |
ENST00000225296.3
|
DHX33
|
DEAH (Asp-Glu-Ala-His) box polypeptide 33 |
| chr3_-_108308241 | 0.05 |
ENST00000295746.8
|
KIAA1524
|
KIAA1524 |
| chr9_-_77643189 | 0.05 |
ENST00000376837.3
|
C9orf41
|
chromosome 9 open reading frame 41 |
| chr5_-_145214893 | 0.05 |
ENST00000394450.2
|
PRELID2
|
PRELI domain containing 2 |
| chr8_+_17354587 | 0.05 |
ENST00000494857.1
ENST00000522656.1 |
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
| chr21_+_38338737 | 0.05 |
ENST00000430068.1
|
AP000704.5
|
AP000704.5 |
| chr16_+_89724188 | 0.05 |
ENST00000301031.4
ENST00000566204.1 ENST00000579310.1 |
SPATA33
|
spermatogenesis associated 33 |
| chr20_-_61493115 | 0.05 |
ENST00000335351.3
ENST00000217162.5 |
TCFL5
|
transcription factor-like 5 (basic helix-loop-helix) |
| chr1_-_43205811 | 0.05 |
ENST00000372539.3
ENST00000296387.1 ENST00000539749.1 |
CLDN19
|
claudin 19 |
| chr1_+_43148625 | 0.05 |
ENST00000436427.1
|
YBX1
|
Y box binding protein 1 |
| chr11_-_64013663 | 0.05 |
ENST00000392210.2
|
PPP1R14B
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr19_-_663147 | 0.05 |
ENST00000606702.1
|
RNF126
|
ring finger protein 126 |
| chr17_+_29158962 | 0.05 |
ENST00000321990.4
|
ATAD5
|
ATPase family, AAA domain containing 5 |
| chr12_-_107487604 | 0.05 |
ENST00000008527.5
|
CRY1
|
cryptochrome 1 (photolyase-like) |
| chr10_+_60145155 | 0.05 |
ENST00000373895.3
|
TFAM
|
transcription factor A, mitochondrial |
| chr12_+_51985001 | 0.05 |
ENST00000354534.6
|
SCN8A
|
sodium channel, voltage gated, type VIII, alpha subunit |
| chr4_-_83351005 | 0.05 |
ENST00000295470.5
|
HNRNPDL
|
heterogeneous nuclear ribonucleoprotein D-like |
| chr20_-_58508702 | 0.05 |
ENST00000357552.3
ENST00000425931.1 |
SYCP2
|
synaptonemal complex protein 2 |
| chr3_-_136471204 | 0.05 |
ENST00000480733.1
ENST00000383202.2 ENST00000236698.5 ENST00000434713.2 |
STAG1
|
stromal antigen 1 |
| chr14_+_64970662 | 0.05 |
ENST00000556965.1
ENST00000554015.1 |
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr6_-_109703600 | 0.05 |
ENST00000512821.1
|
CD164
|
CD164 molecule, sialomucin |
| chrX_+_135579238 | 0.05 |
ENST00000535601.1
ENST00000448450.1 ENST00000425695.1 |
HTATSF1
|
HIV-1 Tat specific factor 1 |
| chr14_+_59951161 | 0.05 |
ENST00000261247.9
ENST00000425728.2 ENST00000556985.1 ENST00000554271.1 ENST00000554795.1 |
JKAMP
|
JNK1/MAPK8-associated membrane protein |
| chr1_+_15480197 | 0.05 |
ENST00000400796.3
ENST00000434578.2 ENST00000376008.2 |
TMEM51
|
transmembrane protein 51 |
| chr3_-_33759541 | 0.05 |
ENST00000468888.2
|
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr6_+_87865262 | 0.04 |
ENST00000369577.3
ENST00000518845.1 ENST00000339907.4 ENST00000496806.2 |
ZNF292
|
zinc finger protein 292 |
| chr3_-_185542761 | 0.04 |
ENST00000457616.2
ENST00000346192.3 |
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr2_+_191513959 | 0.04 |
ENST00000337386.5
ENST00000357215.5 |
NAB1
|
NGFI-A binding protein 1 (EGR1 binding protein 1) |
| chr11_-_117747434 | 0.04 |
ENST00000529335.2
ENST00000530956.1 ENST00000260282.4 |
FXYD6
|
FXYD domain containing ion transport regulator 6 |
| chr6_+_64282447 | 0.04 |
ENST00000370650.2
ENST00000578299.1 |
PTP4A1
|
protein tyrosine phosphatase type IVA, member 1 |
| chr14_-_51411194 | 0.04 |
ENST00000544180.2
|
PYGL
|
phosphorylase, glycogen, liver |
| chr14_-_54955376 | 0.04 |
ENST00000553333.1
|
GMFB
|
glia maturation factor, beta |
| chr14_-_51411146 | 0.04 |
ENST00000532462.1
|
PYGL
|
phosphorylase, glycogen, liver |
| chr15_-_75743915 | 0.04 |
ENST00000394949.4
|
SIN3A
|
SIN3 transcription regulator family member A |
| chr14_-_54955721 | 0.04 |
ENST00000554908.1
|
GMFB
|
glia maturation factor, beta |
| chr10_-_14880002 | 0.04 |
ENST00000465530.1
|
CDNF
|
cerebral dopamine neurotrophic factor |
| chr3_-_15469045 | 0.04 |
ENST00000450816.2
|
METTL6
|
methyltransferase like 6 |
| chr2_+_48667898 | 0.04 |
ENST00000281394.4
ENST00000294952.8 |
PPP1R21
|
protein phosphatase 1, regulatory subunit 21 |
| chr7_+_66093908 | 0.04 |
ENST00000443322.1
ENST00000449064.1 |
KCTD7
|
potassium channel tetramerization domain containing 7 |
| chr12_-_96184533 | 0.04 |
ENST00000343702.4
ENST00000344911.4 |
NTN4
|
netrin 4 |
| chr5_-_2751762 | 0.04 |
ENST00000302057.5
ENST00000382611.6 |
IRX2
|
iroquois homeobox 2 |
| chr19_+_45971246 | 0.04 |
ENST00000585836.1
ENST00000417353.2 ENST00000353609.3 ENST00000591858.1 ENST00000443841.2 ENST00000590335.1 |
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B |
| chr3_-_72496035 | 0.04 |
ENST00000477973.2
|
RYBP
|
RING1 and YY1 binding protein |
| chr15_-_33486897 | 0.04 |
ENST00000320930.7
|
FMN1
|
formin 1 |
| chr15_-_41408339 | 0.04 |
ENST00000401393.3
|
INO80
|
INO80 complex subunit |
| chr4_-_83719983 | 0.04 |
ENST00000319540.4
|
SCD5
|
stearoyl-CoA desaturase 5 |
| chr2_+_37311645 | 0.04 |
ENST00000281932.5
|
GPATCH11
|
G patch domain containing 11 |
| chr10_-_131762105 | 0.04 |
ENST00000368648.3
ENST00000355311.5 |
EBF3
|
early B-cell factor 3 |
| chr7_+_116312411 | 0.04 |
ENST00000456159.1
ENST00000397752.3 ENST00000318493.6 |
MET
|
met proto-oncogene |
| chr3_+_111578640 | 0.04 |
ENST00000393925.3
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2 |
| chr3_+_108308513 | 0.04 |
ENST00000361582.3
|
DZIP3
|
DAZ interacting zinc finger protein 3 |
| chr8_-_101964265 | 0.04 |
ENST00000395958.2
|
YWHAZ
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr11_+_133938820 | 0.04 |
ENST00000299106.4
ENST00000529443.2 |
JAM3
|
junctional adhesion molecule 3 |
| chr2_-_153032484 | 0.04 |
ENST00000263904.4
|
STAM2
|
signal transducing adaptor molecule (SH3 domain and ITAM motif) 2 |
| chr14_+_59655369 | 0.04 |
ENST00000360909.3
ENST00000351081.1 ENST00000556135.1 |
DAAM1
|
dishevelled associated activator of morphogenesis 1 |
| chr3_-_71114066 | 0.04 |
ENST00000485326.2
|
FOXP1
|
forkhead box P1 |
| chr2_+_191513789 | 0.04 |
ENST00000409581.1
|
NAB1
|
NGFI-A binding protein 1 (EGR1 binding protein 1) |
| chr3_+_111578131 | 0.04 |
ENST00000498699.1
|
PHLDB2
|
pleckstrin homology-like domain, family B, member 2 |
| chr7_-_51384451 | 0.04 |
ENST00000441453.1
ENST00000265136.7 ENST00000395542.2 ENST00000395540.2 |
COBL
|
cordon-bleu WH2 repeat protein |
| chr3_-_100120223 | 0.04 |
ENST00000284320.5
|
TOMM70A
|
translocase of outer mitochondrial membrane 70 homolog A (S. cerevisiae) |
| chr4_+_39184024 | 0.04 |
ENST00000399820.3
ENST00000509560.1 ENST00000512112.1 ENST00000288634.7 ENST00000506503.1 |
WDR19
|
WD repeat domain 19 |
| chr22_+_21369316 | 0.04 |
ENST00000413302.2
ENST00000402329.3 ENST00000336296.2 ENST00000401443.1 ENST00000443995.3 |
P2RX6
|
purinergic receptor P2X, ligand-gated ion channel, 6 |
| chr10_+_70091812 | 0.04 |
ENST00000265866.7
|
HNRNPH3
|
heterogeneous nuclear ribonucleoprotein H3 (2H9) |
| chr3_-_33759699 | 0.04 |
ENST00000399362.4
ENST00000359576.5 ENST00000307312.7 |
CLASP2
|
cytoplasmic linker associated protein 2 |
| chr10_-_101769617 | 0.04 |
ENST00000324109.4
ENST00000342239.3 |
DNMBP
|
dynamin binding protein |
| chr5_+_72861560 | 0.04 |
ENST00000296792.4
ENST00000509005.1 ENST00000543251.1 ENST00000508686.1 ENST00000508491.1 |
UTP15
|
UTP15, U3 small nucleolar ribonucleoprotein, homolog (S. cerevisiae) |
| chr14_-_71276211 | 0.04 |
ENST00000381250.4
ENST00000555993.2 |
MAP3K9
|
mitogen-activated protein kinase kinase kinase 9 |
| chr2_-_242255117 | 0.04 |
ENST00000420451.1
ENST00000417540.1 ENST00000310931.4 |
HDLBP
|
high density lipoprotein binding protein |
| chrX_+_133507389 | 0.04 |
ENST00000370800.4
|
PHF6
|
PHD finger protein 6 |
| chr5_+_443268 | 0.04 |
ENST00000512944.1
|
EXOC3
|
exocyst complex component 3 |
| chr6_-_82462425 | 0.04 |
ENST00000369754.3
ENST00000320172.6 ENST00000369756.3 |
FAM46A
|
family with sequence similarity 46, member A |
| chr1_+_39571026 | 0.04 |
ENST00000524432.1
|
MACF1
|
microtubule-actin crosslinking factor 1 |
| chr3_+_111578027 | 0.04 |
ENST00000431670.2
ENST00000412622.1 |
PHLDB2
|
pleckstrin homology-like domain, family B, member 2 |
| chr9_-_77643307 | 0.04 |
ENST00000376834.3
ENST00000376830.3 |
C9orf41
|
chromosome 9 open reading frame 41 |
| chr16_+_53164956 | 0.04 |
ENST00000563410.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr5_-_139283982 | 0.04 |
ENST00000340391.3
|
NRG2
|
neuregulin 2 |
| chr12_+_13197218 | 0.04 |
ENST00000197268.8
|
KIAA1467
|
KIAA1467 |
| chr8_+_22298578 | 0.04 |
ENST00000240139.5
ENST00000289963.8 ENST00000397775.3 |
PPP3CC
|
protein phosphatase 3, catalytic subunit, gamma isozyme |
| chr6_+_125474992 | 0.03 |
ENST00000528193.1
|
TPD52L1
|
tumor protein D52-like 1 |
| chr8_+_17354617 | 0.03 |
ENST00000470360.1
|
SLC7A2
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 |
| chr19_-_39832563 | 0.03 |
ENST00000599274.1
|
CTC-246B18.10
|
CTC-246B18.10 |
| chr3_-_185542817 | 0.03 |
ENST00000382199.2
|
IGF2BP2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr8_-_74884511 | 0.03 |
ENST00000518127.1
|
TCEB1
|
transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C) |
| chr17_-_58469474 | 0.03 |
ENST00000300896.4
|
USP32
|
ubiquitin specific peptidase 32 |
| chr6_+_126070726 | 0.03 |
ENST00000368364.3
|
HEY2
|
hes-related family bHLH transcription factor with YRPW motif 2 |
| chr16_+_66400533 | 0.03 |
ENST00000341529.3
|
CDH5
|
cadherin 5, type 2 (vascular endothelium) |
| chr8_-_30891078 | 0.03 |
ENST00000339382.2
ENST00000475541.1 |
PURG
|
purine-rich element binding protein G |
| chr17_+_43209967 | 0.03 |
ENST00000431281.1
ENST00000591859.1 |
ACBD4
|
acyl-CoA binding domain containing 4 |
| chr4_+_37978891 | 0.03 |
ENST00000446803.2
|
TBC1D1
|
TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1 |
| chr20_-_17662705 | 0.03 |
ENST00000455029.2
|
RRBP1
|
ribosome binding protein 1 |
| chr17_-_39674668 | 0.03 |
ENST00000393981.3
|
KRT15
|
keratin 15 |
| chr9_-_74525658 | 0.03 |
ENST00000333421.6
|
ABHD17B
|
abhydrolase domain containing 17B |
| chr8_-_59572093 | 0.03 |
ENST00000427130.2
|
NSMAF
|
neutral sphingomyelinase (N-SMase) activation associated factor |
| chr7_-_26904317 | 0.03 |
ENST00000345317.2
|
SKAP2
|
src kinase associated phosphoprotein 2 |
| chr11_-_14380664 | 0.03 |
ENST00000545643.1
ENST00000256196.4 |
RRAS2
|
related RAS viral (r-ras) oncogene homolog 2 |
| chr17_+_5185552 | 0.03 |
ENST00000262477.6
ENST00000408982.2 ENST00000575991.1 ENST00000537505.1 ENST00000546142.2 |
RABEP1
|
rabaptin, RAB GTPase binding effector protein 1 |
| chr6_-_109703634 | 0.03 |
ENST00000324953.5
ENST00000310786.4 ENST00000275080.7 ENST00000413644.2 |
CD164
|
CD164 molecule, sialomucin |
| chrX_+_133507327 | 0.03 |
ENST00000332070.3
ENST00000394292.1 ENST00000370799.1 ENST00000416404.2 |
PHF6
|
PHD finger protein 6 |
| chr2_-_136288740 | 0.03 |
ENST00000264159.6
ENST00000536680.1 |
ZRANB3
|
zinc finger, RAN-binding domain containing 3 |
| chr7_+_26191809 | 0.03 |
ENST00000056233.3
|
NFE2L3
|
nuclear factor, erythroid 2-like 3 |
| chr10_+_70661014 | 0.03 |
ENST00000373585.3
|
DDX50
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 50 |
| chr4_+_95373037 | 0.03 |
ENST00000359265.4
ENST00000437932.1 ENST00000380180.3 ENST00000318007.5 ENST00000450793.1 ENST00000538141.1 ENST00000317968.4 ENST00000512274.1 ENST00000503974.1 ENST00000504489.1 ENST00000542407.1 |
PDLIM5
|
PDZ and LIM domain 5 |
| chr14_+_100705322 | 0.03 |
ENST00000262238.4
|
YY1
|
YY1 transcription factor |
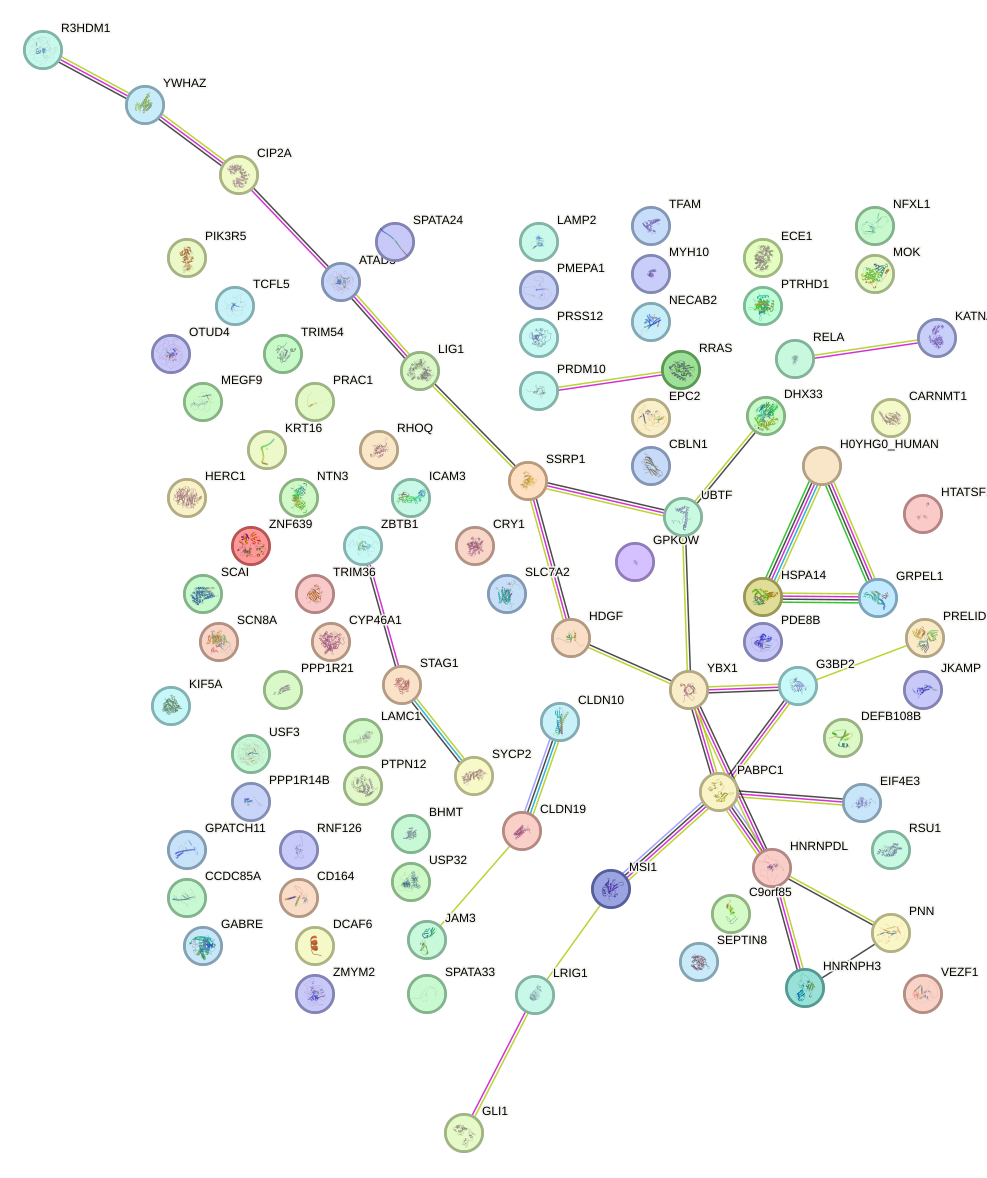
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB7A_ZBTB7C
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0060032 | notochord regression(GO:0060032) |
| 0.0 | 0.1 | GO:0090598 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.0 | 0.1 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 0.0 | 0.1 | GO:0061461 | lysine import(GO:0034226) L-lysine import(GO:0061461) L-lysine import into cell(GO:1903410) |
| 0.0 | 0.0 | GO:0032240 | negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.0 | 0.1 | GO:0051083 | 'de novo' cotranslational protein folding(GO:0051083) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.2 | GO:0021592 | fourth ventricle development(GO:0021592) |
| 0.0 | 0.1 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.0 | 0.2 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.0 | 0.1 | GO:0060168 | positive regulation of adenosine receptor signaling pathway(GO:0060168) regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.0 | 0.1 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.1 | GO:0035937 | estrogen secretion(GO:0035937) estradiol secretion(GO:0035938) regulation of estrogen secretion(GO:2000861) regulation of estradiol secretion(GO:2000864) |
| 0.0 | 0.1 | GO:0006117 | acetaldehyde metabolic process(GO:0006117) |
| 0.0 | 0.1 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.0 | 0.1 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.1 | GO:0034959 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.0 | 0.1 | GO:1990262 | regulation of anti-Mullerian hormone signaling pathway(GO:1902612) negative regulation of anti-Mullerian hormone signaling pathway(GO:1902613) anti-Mullerian hormone signaling pathway(GO:1990262) |
| 0.0 | 0.1 | GO:2000176 | regulation of pro-T cell differentiation(GO:2000174) positive regulation of pro-T cell differentiation(GO:2000176) |
| 0.0 | 0.1 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.0 | 0.2 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.0 | GO:0072081 | proximal/distal pattern formation involved in nephron development(GO:0072047) specification of nephron tubule identity(GO:0072081) pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.0 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.0 | 0.0 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) |
| 0.0 | 0.1 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.1 | GO:0021707 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) positive regulation of long term synaptic depression(GO:1900454) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.4 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.2 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.1 | GO:0097636 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.0 | 0.1 | GO:0035525 | NF-kappaB p50/p65 complex(GO:0035525) |
| 0.0 | 0.5 | GO:0005845 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.1 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.0 | 0.1 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.1 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.6 | GO:0097546 | ciliary base(GO:0097546) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.1 | GO:0008184 | purine nucleobase binding(GO:0002060) glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.1 | GO:0070363 | mitochondrial light strand promoter sense binding(GO:0070363) |
| 0.0 | 0.3 | GO:0000182 | rDNA binding(GO:0000182) |
| 0.0 | 0.1 | GO:0005292 | high-affinity basic amino acid transmembrane transporter activity(GO:0005287) high-affinity arginine transmembrane transporter activity(GO:0005289) high-affinity lysine transmembrane transporter activity(GO:0005292) |
| 0.0 | 0.1 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.0 | 0.1 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.1 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |