Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
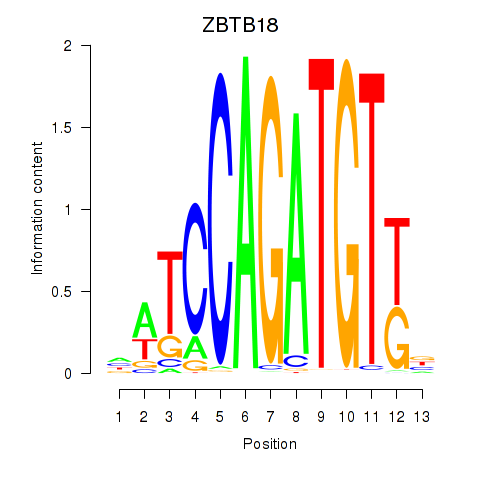
Results for ZBTB18
Z-value: 0.37
Transcription factors associated with ZBTB18
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB18
|
ENSG00000179456.9 | zinc finger and BTB domain containing 18 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZBTB18 | hg19_v2_chr1_+_244214577_244214593 | 0.05 | 9.5e-01 | Click! |
Activity profile of ZBTB18 motif
Sorted Z-values of ZBTB18 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_+_94577074 | 0.26 |
ENST00000444961.1
ENST00000448882.1 ENST00000557098.1 ENST00000554800.1 ENST00000556544.1 ENST00000298902.5 ENST00000555819.1 ENST00000557634.1 ENST00000555744.1 |
IFI27
|
interferon, alpha-inducible protein 27 |
| chr19_+_9203855 | 0.15 |
ENST00000429566.3
|
OR1M1
|
olfactory receptor, family 1, subfamily M, member 1 |
| chr11_+_94227129 | 0.13 |
ENST00000540349.1
ENST00000535502.1 ENST00000545130.1 ENST00000544253.1 ENST00000541144.1 |
ANKRD49
|
ankyrin repeat domain 49 |
| chr1_+_78354175 | 0.13 |
ENST00000401035.3
ENST00000457030.1 ENST00000330010.8 |
NEXN
|
nexilin (F actin binding protein) |
| chr10_-_7829909 | 0.12 |
ENST00000379562.4
ENST00000543003.1 ENST00000535925.1 |
KIN
|
KIN, antigenic determinant of recA protein homolog (mouse) |
| chr3_-_160167301 | 0.12 |
ENST00000494486.1
|
TRIM59
|
tripartite motif containing 59 |
| chrX_-_101771645 | 0.11 |
ENST00000289373.4
|
TMSB15A
|
thymosin beta 15a |
| chrX_+_150151824 | 0.11 |
ENST00000455596.1
ENST00000448905.2 |
HMGB3
|
high mobility group box 3 |
| chr3_-_160167508 | 0.10 |
ENST00000479460.1
|
TRIM59
|
tripartite motif containing 59 |
| chr2_-_65659762 | 0.10 |
ENST00000440972.1
|
SPRED2
|
sprouty-related, EVH1 domain containing 2 |
| chr11_-_94226964 | 0.10 |
ENST00000538923.1
ENST00000540013.1 ENST00000407439.3 ENST00000393241.4 |
MRE11A
|
MRE11 meiotic recombination 11 homolog A (S. cerevisiae) |
| chr15_-_55611306 | 0.09 |
ENST00000563262.1
|
RAB27A
|
RAB27A, member RAS oncogene family |
| chr19_-_10341948 | 0.09 |
ENST00000590320.1
ENST00000592342.1 ENST00000588952.1 |
S1PR2
DNMT1
|
sphingosine-1-phosphate receptor 2 DNA (cytosine-5-)-methyltransferase 1 |
| chr19_+_38880695 | 0.08 |
ENST00000587947.1
ENST00000338502.4 |
SPRED3
|
sprouty-related, EVH1 domain containing 3 |
| chr11_-_94227029 | 0.08 |
ENST00000323977.3
ENST00000536754.1 ENST00000323929.3 |
MRE11A
|
MRE11 meiotic recombination 11 homolog A (S. cerevisiae) |
| chr1_+_78354297 | 0.08 |
ENST00000334785.7
|
NEXN
|
nexilin (F actin binding protein) |
| chrX_+_43515467 | 0.08 |
ENST00000338702.3
ENST00000542639.1 |
MAOA
|
monoamine oxidase A |
| chr19_+_11649532 | 0.08 |
ENST00000252456.2
ENST00000592923.1 ENST00000535659.2 |
CNN1
|
calponin 1, basic, smooth muscle |
| chr19_-_36001386 | 0.07 |
ENST00000461300.1
|
DMKN
|
dermokine |
| chr6_-_163148780 | 0.07 |
ENST00000366892.1
ENST00000366898.1 ENST00000366897.1 ENST00000366896.1 |
PARK2
|
parkin RBR E3 ubiquitin protein ligase |
| chr3_+_100211412 | 0.07 |
ENST00000323523.4
ENST00000403410.1 ENST00000449609.1 |
TMEM45A
|
transmembrane protein 45A |
| chr6_-_153304148 | 0.07 |
ENST00000229758.3
|
FBXO5
|
F-box protein 5 |
| chr1_+_78354243 | 0.07 |
ENST00000294624.8
|
NEXN
|
nexilin (F actin binding protein) |
| chr3_-_145940126 | 0.07 |
ENST00000498625.1
|
PLSCR4
|
phospholipid scramblase 4 |
| chr12_-_68845417 | 0.07 |
ENST00000542875.1
|
RP11-81H14.2
|
RP11-81H14.2 |
| chr15_+_49715449 | 0.06 |
ENST00000560979.1
|
FGF7
|
fibroblast growth factor 7 |
| chr9_-_99329098 | 0.06 |
ENST00000452280.1
|
CDC14B
|
cell division cycle 14B |
| chr14_+_64970662 | 0.06 |
ENST00000556965.1
ENST00000554015.1 |
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr9_-_13165457 | 0.06 |
ENST00000542239.1
ENST00000538841.1 ENST00000433359.2 |
MPDZ
|
multiple PDZ domain protein |
| chr16_+_67563250 | 0.06 |
ENST00000566907.1
|
FAM65A
|
family with sequence similarity 65, member A |
| chr10_+_7830125 | 0.06 |
ENST00000335698.4
ENST00000541227.1 |
ATP5C1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 |
| chr3_+_121311966 | 0.06 |
ENST00000338040.4
|
FBXO40
|
F-box protein 40 |
| chr2_+_201994042 | 0.06 |
ENST00000417748.1
|
CFLAR
|
CASP8 and FADD-like apoptosis regulator |
| chr1_+_78354330 | 0.06 |
ENST00000440324.1
|
NEXN
|
nexilin (F actin binding protein) |
| chr2_-_159237472 | 0.06 |
ENST00000409187.1
|
CCDC148
|
coiled-coil domain containing 148 |
| chr11_-_1606513 | 0.06 |
ENST00000382171.2
|
KRTAP5-1
|
keratin associated protein 5-1 |
| chrX_+_150151752 | 0.06 |
ENST00000325307.7
|
HMGB3
|
high mobility group box 3 |
| chr7_+_105172612 | 0.06 |
ENST00000493041.1
|
RINT1
|
RAD50 interactor 1 |
| chr8_-_49833978 | 0.05 |
ENST00000020945.1
|
SNAI2
|
snail family zinc finger 2 |
| chr1_-_207206092 | 0.05 |
ENST00000359470.5
ENST00000461135.2 |
C1orf116
|
chromosome 1 open reading frame 116 |
| chr8_-_49834299 | 0.05 |
ENST00000396822.1
|
SNAI2
|
snail family zinc finger 2 |
| chr12_-_68845165 | 0.05 |
ENST00000360485.3
ENST00000441255.2 |
RP11-81H14.2
|
RP11-81H14.2 |
| chr5_-_142784101 | 0.05 |
ENST00000503201.1
ENST00000502892.1 |
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr4_+_75480629 | 0.05 |
ENST00000380846.3
|
AREGB
|
amphiregulin B |
| chr19_+_54135310 | 0.05 |
ENST00000376650.1
|
DPRX
|
divergent-paired related homeobox |
| chr15_+_55611401 | 0.05 |
ENST00000566999.1
|
PIGB
|
phosphatidylinositol glycan anchor biosynthesis, class B |
| chr13_-_28545276 | 0.05 |
ENST00000381020.7
|
CDX2
|
caudal type homeobox 2 |
| chr11_-_5537920 | 0.05 |
ENST00000380184.1
|
UBQLNL
|
ubiquilin-like |
| chr1_-_113160826 | 0.04 |
ENST00000538187.1
ENST00000369664.1 |
ST7L
|
suppression of tumorigenicity 7 like |
| chr10_+_7830092 | 0.04 |
ENST00000356708.7
|
ATP5C1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 |
| chrX_+_9880412 | 0.04 |
ENST00000418909.2
|
SHROOM2
|
shroom family member 2 |
| chr5_+_159895275 | 0.04 |
ENST00000517927.1
|
MIR146A
|
microRNA 146a |
| chr1_-_156647189 | 0.04 |
ENST00000368223.3
|
NES
|
nestin |
| chr15_-_57210769 | 0.04 |
ENST00000559000.1
|
ZNF280D
|
zinc finger protein 280D |
| chr4_+_75310851 | 0.04 |
ENST00000395748.3
ENST00000264487.2 |
AREG
|
amphiregulin |
| chr2_-_7005785 | 0.04 |
ENST00000256722.5
ENST00000404168.1 ENST00000458098.1 |
CMPK2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial |
| chr11_-_101454658 | 0.04 |
ENST00000344327.3
|
TRPC6
|
transient receptor potential cation channel, subfamily C, member 6 |
| chr11_+_32154200 | 0.04 |
ENST00000525133.1
|
RP1-65P5.5
|
RP1-65P5.5 |
| chrX_+_99899180 | 0.04 |
ENST00000373004.3
|
SRPX2
|
sushi-repeat containing protein, X-linked 2 |
| chr10_-_105437909 | 0.04 |
ENST00000540321.1
|
SH3PXD2A
|
SH3 and PX domains 2A |
| chrX_-_154563889 | 0.04 |
ENST00000369449.2
ENST00000321926.4 |
CLIC2
|
chloride intracellular channel 2 |
| chr19_-_36001286 | 0.04 |
ENST00000602679.1
ENST00000492341.2 ENST00000472252.2 ENST00000602781.1 ENST00000402589.2 ENST00000458071.1 ENST00000436012.1 ENST00000443640.1 ENST00000450261.1 ENST00000467637.1 ENST00000480502.1 ENST00000474928.1 ENST00000414866.2 ENST00000392206.2 ENST00000488892.1 |
DMKN
|
dermokine |
| chr10_-_104913367 | 0.04 |
ENST00000423468.2
|
NT5C2
|
5'-nucleotidase, cytosolic II |
| chr3_-_44552094 | 0.04 |
ENST00000436261.1
|
ZNF852
|
zinc finger protein 852 |
| chr2_+_152214098 | 0.04 |
ENST00000243347.3
|
TNFAIP6
|
tumor necrosis factor, alpha-induced protein 6 |
| chr8_+_38662960 | 0.04 |
ENST00000524193.1
|
TACC1
|
transforming, acidic coiled-coil containing protein 1 |
| chr17_+_47653178 | 0.04 |
ENST00000328741.5
|
NXPH3
|
neurexophilin 3 |
| chr1_-_151882031 | 0.03 |
ENST00000489410.1
|
THEM4
|
thioesterase superfamily member 4 |
| chr11_+_32112431 | 0.03 |
ENST00000054950.3
|
RCN1
|
reticulocalbin 1, EF-hand calcium binding domain |
| chr7_-_135433460 | 0.03 |
ENST00000415751.1
|
FAM180A
|
family with sequence similarity 180, member A |
| chr2_-_152146385 | 0.03 |
ENST00000414946.1
ENST00000243346.5 |
NMI
|
N-myc (and STAT) interactor |
| chr3_-_160167540 | 0.03 |
ENST00000496222.1
ENST00000471396.1 ENST00000471155.1 ENST00000309784.4 |
TRIM59
|
tripartite motif containing 59 |
| chr7_-_154863264 | 0.03 |
ENST00000395731.2
ENST00000543018.1 |
HTR5A-AS1
|
HTR5A antisense RNA 1 |
| chr9_+_72658490 | 0.03 |
ENST00000377182.4
|
MAMDC2
|
MAM domain containing 2 |
| chr11_-_6008215 | 0.03 |
ENST00000332249.4
|
OR52L1
|
olfactory receptor, family 52, subfamily L, member 1 |
| chr8_+_1772132 | 0.03 |
ENST00000349830.3
ENST00000520359.1 ENST00000518288.1 ENST00000398560.1 |
ARHGEF10
|
Rho guanine nucleotide exchange factor (GEF) 10 |
| chr11_-_119252359 | 0.03 |
ENST00000455332.2
|
USP2
|
ubiquitin specific peptidase 2 |
| chr1_-_2458026 | 0.03 |
ENST00000435556.3
ENST00000378466.3 |
PANK4
|
pantothenate kinase 4 |
| chr1_+_19967014 | 0.03 |
ENST00000428975.1
|
NBL1
|
neuroblastoma 1, DAN family BMP antagonist |
| chr1_+_53527854 | 0.03 |
ENST00000371500.3
ENST00000395871.2 ENST00000312553.5 |
PODN
|
podocan |
| chr9_+_100174232 | 0.03 |
ENST00000355295.4
|
TDRD7
|
tudor domain containing 7 |
| chr16_-_18470696 | 0.03 |
ENST00000427999.2
|
RP11-1212A22.4
|
LOC339047 protein; Nuclear pore complex-interacting protein family member A3; Nuclear pore complex-interacting protein family member A5; Protein PKD1P1 |
| chr15_+_92937144 | 0.03 |
ENST00000539113.1
ENST00000555434.1 |
ST8SIA2
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 |
| chr1_-_224517823 | 0.03 |
ENST00000469968.1
ENST00000436927.1 ENST00000469075.1 ENST00000488718.1 ENST00000482491.1 ENST00000340871.4 ENST00000492281.1 ENST00000361463.3 ENST00000391875.2 ENST00000461546.1 |
NVL
|
nuclear VCP-like |
| chr14_+_67707826 | 0.03 |
ENST00000261681.4
|
MPP5
|
membrane protein, palmitoylated 5 (MAGUK p55 subfamily member 5) |
| chr3_+_49840685 | 0.03 |
ENST00000333323.4
|
FAM212A
|
family with sequence similarity 212, member A |
| chr4_-_87855851 | 0.03 |
ENST00000473559.1
|
C4orf36
|
chromosome 4 open reading frame 36 |
| chr4_+_75311019 | 0.03 |
ENST00000502307.1
|
AREG
|
amphiregulin |
| chr17_+_47653471 | 0.03 |
ENST00000513748.1
|
NXPH3
|
neurexophilin 3 |
| chr3_+_183894737 | 0.03 |
ENST00000432591.1
ENST00000431779.1 |
AP2M1
|
adaptor-related protein complex 2, mu 1 subunit |
| chr5_-_151304337 | 0.03 |
ENST00000455880.2
ENST00000545569.1 ENST00000274576.4 |
GLRA1
|
glycine receptor, alpha 1 |
| chr5_-_180552304 | 0.03 |
ENST00000329365.2
|
OR2V1
|
olfactory receptor, family 2, subfamily V, member 1 |
| chr12_-_11175219 | 0.03 |
ENST00000390673.2
|
TAS2R19
|
taste receptor, type 2, member 19 |
| chr1_-_168698433 | 0.03 |
ENST00000367817.3
|
DPT
|
dermatopontin |
| chr14_+_64971292 | 0.02 |
ENST00000358738.3
ENST00000394712.2 |
ZBTB1
|
zinc finger and BTB domain containing 1 |
| chr9_+_273038 | 0.02 |
ENST00000487230.1
ENST00000469391.1 |
DOCK8
|
dedicator of cytokinesis 8 |
| chrX_+_9880590 | 0.02 |
ENST00000452575.1
|
SHROOM2
|
shroom family member 2 |
| chr3_-_105588231 | 0.02 |
ENST00000545639.1
ENST00000394027.3 ENST00000438603.1 ENST00000447441.1 ENST00000443752.1 |
CBLB
|
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr4_+_47487285 | 0.02 |
ENST00000273859.3
ENST00000504445.1 |
ATP10D
|
ATPase, class V, type 10D |
| chr19_+_50084561 | 0.02 |
ENST00000246794.5
|
PRRG2
|
proline rich Gla (G-carboxyglutamic acid) 2 |
| chr9_+_2029019 | 0.02 |
ENST00000382194.1
|
SMARCA2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr3_-_37216055 | 0.02 |
ENST00000336686.4
|
LRRFIP2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr2_+_191045562 | 0.02 |
ENST00000340623.4
|
C2orf88
|
chromosome 2 open reading frame 88 |
| chr12_+_53846612 | 0.02 |
ENST00000551104.1
|
PCBP2
|
poly(rC) binding protein 2 |
| chr12_+_27619743 | 0.02 |
ENST00000298876.4
ENST00000416383.1 |
SMCO2
|
single-pass membrane protein with coiled-coil domains 2 |
| chr12_+_112563335 | 0.02 |
ENST00000549358.1
ENST00000257604.5 ENST00000548092.1 ENST00000552896.1 |
TRAFD1
|
TRAF-type zinc finger domain containing 1 |
| chr12_+_57854274 | 0.02 |
ENST00000528432.1
|
GLI1
|
GLI family zinc finger 1 |
| chr2_+_219247021 | 0.02 |
ENST00000539932.1
|
SLC11A1
|
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 1 |
| chr2_+_189839046 | 0.02 |
ENST00000304636.3
ENST00000317840.5 |
COL3A1
|
collagen, type III, alpha 1 |
| chr3_+_124303539 | 0.02 |
ENST00000428018.2
|
KALRN
|
kalirin, RhoGEF kinase |
| chr5_-_142784003 | 0.02 |
ENST00000416954.2
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr12_-_64616019 | 0.02 |
ENST00000311915.8
ENST00000398055.3 ENST00000544871.1 |
C12orf66
|
chromosome 12 open reading frame 66 |
| chr15_+_55611128 | 0.02 |
ENST00000164305.5
ENST00000539642.1 |
PIGB
|
phosphatidylinositol glycan anchor biosynthesis, class B |
| chr12_-_104443890 | 0.02 |
ENST00000547583.1
ENST00000360814.4 ENST00000546851.1 |
GLT8D2
|
glycosyltransferase 8 domain containing 2 |
| chr6_-_16761678 | 0.02 |
ENST00000244769.4
ENST00000436367.1 |
ATXN1
|
ataxin 1 |
| chr6_-_30523865 | 0.02 |
ENST00000433809.1
|
GNL1
|
guanine nucleotide binding protein-like 1 |
| chr9_+_100174344 | 0.02 |
ENST00000422139.2
|
TDRD7
|
tudor domain containing 7 |
| chrX_-_153141434 | 0.02 |
ENST00000407935.2
ENST00000439496.1 |
L1CAM
|
L1 cell adhesion molecule |
| chr16_-_49698136 | 0.02 |
ENST00000535559.1
|
ZNF423
|
zinc finger protein 423 |
| chr15_+_57210961 | 0.02 |
ENST00000557843.1
|
TCF12
|
transcription factor 12 |
| chr16_-_30394143 | 0.02 |
ENST00000321367.3
ENST00000571393.1 |
SEPT1
|
septin 1 |
| chr12_+_26126681 | 0.02 |
ENST00000542865.1
|
RASSF8
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr1_-_92951607 | 0.02 |
ENST00000427103.1
|
GFI1
|
growth factor independent 1 transcription repressor |
| chr6_+_163148973 | 0.02 |
ENST00000366888.2
|
PACRG
|
PARK2 co-regulated |
| chr3_-_145940214 | 0.02 |
ENST00000481701.1
|
PLSCR4
|
phospholipid scramblase 4 |
| chr16_-_30393752 | 0.02 |
ENST00000566517.1
ENST00000605106.1 |
SEPT1
SEPT1
|
septin 1 Uncharacterized protein |
| chr17_-_62050278 | 0.02 |
ENST00000578147.1
ENST00000435607.1 |
SCN4A
|
sodium channel, voltage-gated, type IV, alpha subunit |
| chr15_+_57210818 | 0.02 |
ENST00000438423.2
ENST00000267811.5 ENST00000452095.2 ENST00000559609.1 ENST00000333725.5 |
TCF12
|
transcription factor 12 |
| chrX_-_48056199 | 0.01 |
ENST00000311798.1
ENST00000347757.1 |
SSX5
|
synovial sarcoma, X breakpoint 5 |
| chr9_+_127023704 | 0.01 |
ENST00000373596.1
ENST00000425237.1 |
NEK6
|
NIMA-related kinase 6 |
| chr10_+_97733786 | 0.01 |
ENST00000371198.2
|
CC2D2B
|
coiled-coil and C2 domain containing 2B |
| chrX_+_153770421 | 0.01 |
ENST00000369609.5
ENST00000369607.1 |
IKBKG
|
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr5_-_142784888 | 0.01 |
ENST00000514699.1
|
NR3C1
|
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr11_-_119252425 | 0.01 |
ENST00000260187.2
|
USP2
|
ubiquitin specific peptidase 2 |
| chrX_-_19688475 | 0.01 |
ENST00000541422.1
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chr11_+_59480899 | 0.01 |
ENST00000300150.7
|
STX3
|
syntaxin 3 |
| chr17_+_64961026 | 0.01 |
ENST00000262138.3
|
CACNG4
|
calcium channel, voltage-dependent, gamma subunit 4 |
| chr2_-_145278475 | 0.01 |
ENST00000558170.2
|
ZEB2
|
zinc finger E-box binding homeobox 2 |
| chrX_-_30877837 | 0.01 |
ENST00000378930.3
|
TAB3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chr13_+_97874574 | 0.01 |
ENST00000343600.4
ENST00000345429.6 ENST00000376673.3 |
MBNL2
|
muscleblind-like splicing regulator 2 |
| chr10_-_48050538 | 0.01 |
ENST00000420079.2
|
ASAH2C
|
N-acylsphingosine amidohydrolase (non-lysosomal ceramidase) 2C |
| chr3_+_124303512 | 0.01 |
ENST00000454902.1
|
KALRN
|
kalirin, RhoGEF kinase |
| chr8_-_29592736 | 0.01 |
ENST00000518623.1
|
LINC00589
|
long intergenic non-protein coding RNA 589 |
| chr4_+_156824840 | 0.01 |
ENST00000536354.2
|
TDO2
|
tryptophan 2,3-dioxygenase |
| chr2_-_225434538 | 0.01 |
ENST00000409096.1
|
CUL3
|
cullin 3 |
| chr5_-_180688105 | 0.01 |
ENST00000327767.4
|
TRIM52
|
tripartite motif containing 52 |
| chr4_-_100009856 | 0.01 |
ENST00000296412.8
|
ADH5
|
alcohol dehydrogenase 5 (class III), chi polypeptide |
| chr7_-_47578840 | 0.01 |
ENST00000450444.1
|
TNS3
|
tensin 3 |
| chr2_+_33359473 | 0.01 |
ENST00000432635.1
|
LTBP1
|
latent transforming growth factor beta binding protein 1 |
| chr6_-_74363803 | 0.01 |
ENST00000355773.5
|
SLC17A5
|
solute carrier family 17 (acidic sugar transporter), member 5 |
| chr8_-_17555164 | 0.01 |
ENST00000297488.6
|
MTUS1
|
microtubule associated tumor suppressor 1 |
| chr13_+_113699029 | 0.01 |
ENST00000423251.1
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like |
| chr12_-_48119340 | 0.01 |
ENST00000229003.3
|
ENDOU
|
endonuclease, polyU-specific |
| chr8_+_21823726 | 0.01 |
ENST00000433566.4
|
XPO7
|
exportin 7 |
| chr2_+_177053307 | 0.01 |
ENST00000331462.4
|
HOXD1
|
homeobox D1 |
| chr17_-_15168624 | 0.00 |
ENST00000312280.3
ENST00000494511.1 ENST00000580584.1 |
PMP22
|
peripheral myelin protein 22 |
| chr22_+_42372931 | 0.00 |
ENST00000328414.8
ENST00000396425.3 |
SEPT3
|
septin 3 |
| chr2_-_163100045 | 0.00 |
ENST00000188790.4
|
FAP
|
fibroblast activation protein, alpha |
| chr17_+_32582293 | 0.00 |
ENST00000580907.1
ENST00000225831.4 |
CCL2
|
chemokine (C-C motif) ligand 2 |
| chr4_-_152149033 | 0.00 |
ENST00000514152.1
|
SH3D19
|
SH3 domain containing 19 |
| chr8_-_11058847 | 0.00 |
ENST00000297303.4
ENST00000416569.2 |
XKR6
|
XK, Kell blood group complex subunit-related family, member 6 |
| chr10_-_95242044 | 0.00 |
ENST00000371501.4
ENST00000371502.4 ENST00000371489.1 |
MYOF
|
myoferlin |
| chr12_+_112563303 | 0.00 |
ENST00000412615.2
|
TRAFD1
|
TRAF-type zinc finger domain containing 1 |
| chr10_+_105253661 | 0.00 |
ENST00000369780.4
|
NEURL
|
neuralized E3 ubiquitin protein ligase 1 |
| chr11_-_34379546 | 0.00 |
ENST00000435224.2
|
ABTB2
|
ankyrin repeat and BTB (POZ) domain containing 2 |
| chr5_+_118690466 | 0.00 |
ENST00000503646.1
|
TNFAIP8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr17_+_32907768 | 0.00 |
ENST00000321639.5
|
TMEM132E
|
transmembrane protein 132E |
| chr10_-_95241951 | 0.00 |
ENST00000358334.5
ENST00000359263.4 ENST00000371488.3 |
MYOF
|
myoferlin |
| chr7_-_135433534 | 0.00 |
ENST00000338588.3
|
FAM180A
|
family with sequence similarity 180, member A |
| chr19_-_36001113 | 0.00 |
ENST00000434389.1
|
DMKN
|
dermokine |
| chr2_-_163099885 | 0.00 |
ENST00000443424.1
|
FAP
|
fibroblast activation protein, alpha |
| chr17_+_19912640 | 0.00 |
ENST00000395527.4
ENST00000583482.2 ENST00000583528.1 ENST00000583463.1 |
SPECC1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr15_+_49715293 | 0.00 |
ENST00000267843.4
ENST00000560270.1 |
FGF7
|
fibroblast growth factor 7 |
| chr14_-_61748550 | 0.00 |
ENST00000555868.1
|
TMEM30B
|
transmembrane protein 30B |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB18
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.1 | GO:0007057 | spindle assembly involved in female meiosis I(GO:0007057) |
| 0.0 | 0.1 | GO:0008057 | eye pigment granule organization(GO:0008057) |
| 0.0 | 0.1 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.1 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.3 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.1 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.0 | 0.1 | GO:0032776 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.0 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.0 | GO:0070939 | Dsl1p complex(GO:0070939) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.0 | GO:0050146 | nucleoside phosphotransferase activity(GO:0050146) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.0 | GO:0009041 | uridylate kinase activity(GO:0009041) |