Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
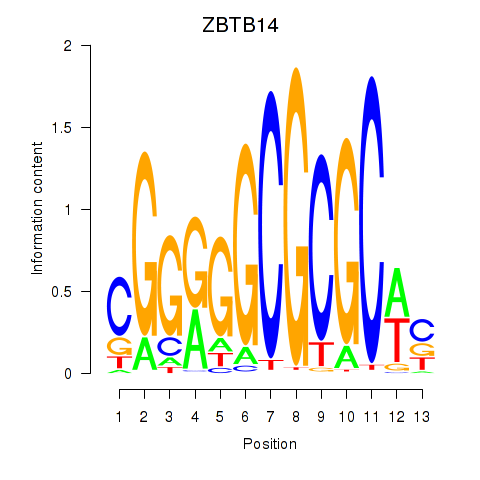
Results for ZBTB14
Z-value: 0.41
Transcription factors associated with ZBTB14
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ZBTB14
|
ENSG00000198081.6 | zinc finger and BTB domain containing 14 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ZBTB14 | hg19_v2_chr18_-_5296001_5296045 | 0.91 | 8.9e-02 | Click! |
Activity profile of ZBTB14 motif
Sorted Z-values of ZBTB14 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_176560595 | 0.22 |
ENST00000508896.1
|
NSD1
|
nuclear receptor binding SET domain protein 1 |
| chr14_+_60716159 | 0.11 |
ENST00000325658.3
|
PPM1A
|
protein phosphatase, Mg2+/Mn2+ dependent, 1A |
| chr9_+_103189536 | 0.10 |
ENST00000374885.1
|
MSANTD3
|
Myb/SANT-like DNA-binding domain containing 3 |
| chr9_+_103189660 | 0.10 |
ENST00000374886.3
|
MSANTD3
|
Myb/SANT-like DNA-binding domain containing 3 |
| chr22_+_24129138 | 0.09 |
ENST00000417137.1
ENST00000344921.6 ENST00000263121.7 ENST00000407422.3 ENST00000407082.3 |
SMARCB1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1 |
| chr19_+_1407733 | 0.09 |
ENST00000592453.1
|
DAZAP1
|
DAZ associated protein 1 |
| chr5_+_98264867 | 0.09 |
ENST00000513175.1
|
CTD-2007H13.3
|
CTD-2007H13.3 |
| chr22_+_29999647 | 0.09 |
ENST00000334961.7
ENST00000353887.4 |
NF2
|
neurofibromin 2 (merlin) |
| chr1_-_33336414 | 0.09 |
ENST00000373471.3
ENST00000609187.1 |
FNDC5
|
fibronectin type III domain containing 5 |
| chr19_+_35521699 | 0.09 |
ENST00000415950.3
|
SCN1B
|
sodium channel, voltage-gated, type I, beta subunit |
| chr12_+_1100423 | 0.08 |
ENST00000592048.1
|
ERC1
|
ELKS/RAB6-interacting/CAST family member 1 |
| chr9_+_103189458 | 0.08 |
ENST00000398977.2
|
MSANTD3
|
Myb/SANT-like DNA-binding domain containing 3 |
| chr2_-_28113217 | 0.08 |
ENST00000444339.2
|
RBKS
|
ribokinase |
| chr14_+_74058410 | 0.07 |
ENST00000326303.4
|
ACOT4
|
acyl-CoA thioesterase 4 |
| chr20_+_57466629 | 0.07 |
ENST00000371081.1
ENST00000338783.6 |
GNAS
|
GNAS complex locus |
| chr7_+_74072288 | 0.07 |
ENST00000443166.1
|
GTF2I
|
general transcription factor IIi |
| chr1_-_40367530 | 0.07 |
ENST00000372816.2
ENST00000372815.1 |
MYCL
|
v-myc avian myelocytomatosis viral oncogene lung carcinoma derived homolog |
| chr2_-_55646957 | 0.07 |
ENST00000263630.8
|
CCDC88A
|
coiled-coil domain containing 88A |
| chr20_-_62462566 | 0.07 |
ENST00000245663.4
ENST00000302995.2 |
ZBTB46
|
zinc finger and BTB domain containing 46 |
| chr11_-_64889252 | 0.07 |
ENST00000525297.1
ENST00000529259.1 |
FAU
|
Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed |
| chr3_-_49449350 | 0.07 |
ENST00000454011.2
ENST00000445425.1 ENST00000422781.1 |
RHOA
|
ras homolog family member A |
| chr2_-_17935027 | 0.06 |
ENST00000446852.1
|
SMC6
|
structural maintenance of chromosomes 6 |
| chr11_-_125932685 | 0.06 |
ENST00000527967.1
|
CDON
|
cell adhesion associated, oncogene regulated |
| chr19_+_35521616 | 0.06 |
ENST00000595652.1
|
SCN1B
|
sodium channel, voltage-gated, type I, beta subunit |
| chr7_+_79764104 | 0.06 |
ENST00000351004.3
|
GNAI1
|
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
| chr14_-_104313824 | 0.06 |
ENST00000553739.1
ENST00000202556.9 |
PPP1R13B
|
protein phosphatase 1, regulatory subunit 13B |
| chr1_+_178694408 | 0.06 |
ENST00000324778.5
|
RALGPS2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr10_+_21823079 | 0.06 |
ENST00000377100.3
ENST00000377072.3 ENST00000446906.2 |
MLLT10
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 |
| chr12_-_124457163 | 0.06 |
ENST00000535556.1
|
CCDC92
|
coiled-coil domain containing 92 |
| chr2_-_55647057 | 0.06 |
ENST00000436346.1
|
CCDC88A
|
coiled-coil domain containing 88A |
| chr13_-_52027134 | 0.06 |
ENST00000311234.4
ENST00000425000.1 ENST00000463928.1 ENST00000442263.3 ENST00000398119.2 |
INTS6
|
integrator complex subunit 6 |
| chr17_+_59477233 | 0.06 |
ENST00000240328.3
|
TBX2
|
T-box 2 |
| chr11_-_129872672 | 0.06 |
ENST00000531431.1
ENST00000527581.1 |
PRDM10
|
PR domain containing 10 |
| chr19_-_13068012 | 0.06 |
ENST00000316939.1
|
GADD45GIP1
|
growth arrest and DNA-damage-inducible, gamma interacting protein 1 |
| chr21_-_46237959 | 0.06 |
ENST00000397898.3
ENST00000411651.2 |
SUMO3
|
small ubiquitin-like modifier 3 |
| chrX_-_153095813 | 0.06 |
ENST00000544474.1
|
PDZD4
|
PDZ domain containing 4 |
| chr17_+_75137460 | 0.06 |
ENST00000587820.1
|
SEC14L1
|
SEC14-like 1 (S. cerevisiae) |
| chr12_-_129308041 | 0.05 |
ENST00000376740.4
|
SLC15A4
|
solute carrier family 15 (oligopeptide transporter), member 4 |
| chr11_-_72852320 | 0.05 |
ENST00000422375.1
|
FCHSD2
|
FCH and double SH3 domains 2 |
| chr20_+_9049742 | 0.05 |
ENST00000437503.1
|
PLCB4
|
phospholipase C, beta 4 |
| chr9_+_103189405 | 0.05 |
ENST00000395067.2
|
MSANTD3
|
Myb/SANT-like DNA-binding domain containing 3 |
| chr8_+_110551925 | 0.05 |
ENST00000395785.2
|
EBAG9
|
estrogen receptor binding site associated, antigen, 9 |
| chr6_-_111136299 | 0.05 |
ENST00000457688.1
|
CDK19
|
cyclin-dependent kinase 19 |
| chr1_-_29508499 | 0.05 |
ENST00000373795.4
|
SRSF4
|
serine/arginine-rich splicing factor 4 |
| chr5_-_178054105 | 0.05 |
ENST00000316308.4
|
CLK4
|
CDC-like kinase 4 |
| chr6_+_18155560 | 0.05 |
ENST00000546309.2
ENST00000388870.2 ENST00000397244.1 |
KDM1B
|
lysine (K)-specific demethylase 1B |
| chr10_+_70587279 | 0.05 |
ENST00000298596.6
ENST00000399169.4 ENST00000399165.4 ENST00000399162.2 |
STOX1
|
storkhead box 1 |
| chr1_+_6845578 | 0.05 |
ENST00000467404.2
ENST00000439411.2 |
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr18_-_12884150 | 0.05 |
ENST00000591115.1
ENST00000309660.5 |
PTPN2
|
protein tyrosine phosphatase, non-receptor type 2 |
| chr11_-_46142615 | 0.05 |
ENST00000529734.1
ENST00000323180.6 |
PHF21A
|
PHD finger protein 21A |
| chr15_-_77712477 | 0.05 |
ENST00000560626.2
|
PEAK1
|
pseudopodium-enriched atypical kinase 1 |
| chr3_+_14989186 | 0.05 |
ENST00000435454.1
ENST00000323373.6 |
NR2C2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr10_+_26986582 | 0.05 |
ENST00000376215.5
ENST00000376203.5 |
PDSS1
|
prenyl (decaprenyl) diphosphate synthase, subunit 1 |
| chr12_+_64798095 | 0.05 |
ENST00000332707.5
|
XPOT
|
exportin, tRNA |
| chr2_+_204193129 | 0.05 |
ENST00000417864.1
|
ABI2
|
abl-interactor 2 |
| chr7_-_155089251 | 0.05 |
ENST00000609974.1
|
AC144652.1
|
AC144652.1 |
| chr3_-_150481218 | 0.05 |
ENST00000482706.1
|
SIAH2
|
siah E3 ubiquitin protein ligase 2 |
| chr6_-_143266297 | 0.05 |
ENST00000367603.2
|
HIVEP2
|
human immunodeficiency virus type I enhancer binding protein 2 |
| chr18_-_12884259 | 0.05 |
ENST00000353319.4
ENST00000327283.3 |
PTPN2
|
protein tyrosine phosphatase, non-receptor type 2 |
| chr4_+_56212505 | 0.05 |
ENST00000505210.1
|
SRD5A3
|
steroid 5 alpha-reductase 3 |
| chr12_-_57824561 | 0.05 |
ENST00000448732.1
|
R3HDM2
|
R3H domain containing 2 |
| chr19_-_40732594 | 0.05 |
ENST00000430325.2
ENST00000433940.1 |
CNTD2
|
cyclin N-terminal domain containing 2 |
| chr17_+_44668035 | 0.05 |
ENST00000398238.4
ENST00000225282.8 |
NSF
|
N-ethylmaleimide-sensitive factor |
| chr4_-_103748271 | 0.05 |
ENST00000343106.5
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr12_-_112450915 | 0.05 |
ENST00000437003.2
ENST00000552374.2 ENST00000550831.3 ENST00000354825.3 ENST00000549537.2 ENST00000355445.3 |
TMEM116
|
transmembrane protein 116 |
| chr2_-_99757876 | 0.05 |
ENST00000539964.1
ENST00000393482.3 |
TSGA10
|
testis specific, 10 |
| chr16_+_90086002 | 0.05 |
ENST00000563936.1
ENST00000536122.1 ENST00000561675.1 |
GAS8
|
growth arrest-specific 8 |
| chrX_-_153363125 | 0.05 |
ENST00000407218.1
ENST00000453960.2 |
MECP2
|
methyl CpG binding protein 2 (Rett syndrome) |
| chr17_+_12693001 | 0.05 |
ENST00000262444.9
|
ARHGAP44
|
Rho GTPase activating protein 44 |
| chr1_+_178694362 | 0.05 |
ENST00000367634.2
|
RALGPS2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr5_+_115177178 | 0.05 |
ENST00000316788.7
|
AP3S1
|
adaptor-related protein complex 3, sigma 1 subunit |
| chr5_+_61602236 | 0.05 |
ENST00000514082.1
ENST00000407818.3 |
KIF2A
|
kinesin heavy chain member 2A |
| chr17_+_35766965 | 0.05 |
ENST00000394395.2
ENST00000589153.1 ENST00000586023.1 |
TADA2A
|
transcriptional adaptor 2A |
| chr6_+_18155632 | 0.05 |
ENST00000297792.5
|
KDM1B
|
lysine (K)-specific demethylase 1B |
| chr2_+_121010413 | 0.04 |
ENST00000404963.3
|
RALB
|
v-ral simian leukemia viral oncogene homolog B |
| chr2_+_17935119 | 0.04 |
ENST00000317402.7
|
GEN1
|
GEN1 Holliday junction 5' flap endonuclease |
| chr9_+_114659046 | 0.04 |
ENST00000374279.3
|
UGCG
|
UDP-glucose ceramide glucosyltransferase |
| chr7_-_32931623 | 0.04 |
ENST00000452926.1
|
KBTBD2
|
kelch repeat and BTB (POZ) domain containing 2 |
| chr11_-_61196858 | 0.04 |
ENST00000413184.2
|
CPSF7
|
cleavage and polyadenylation specific factor 7, 59kDa |
| chr3_+_170136642 | 0.04 |
ENST00000064724.3
ENST00000486975.1 |
CLDN11
|
claudin 11 |
| chr4_-_103748696 | 0.04 |
ENST00000321805.7
|
UBE2D3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr16_+_2521500 | 0.04 |
ENST00000293973.1
|
NTN3
|
netrin 3 |
| chr2_-_17935059 | 0.04 |
ENST00000448223.2
ENST00000381272.4 ENST00000351948.4 |
SMC6
|
structural maintenance of chromosomes 6 |
| chr9_+_127020503 | 0.04 |
ENST00000545174.1
ENST00000444973.1 ENST00000454453.1 |
NEK6
|
NIMA-related kinase 6 |
| chr8_+_66556936 | 0.04 |
ENST00000262146.4
|
MTFR1
|
mitochondrial fission regulator 1 |
| chr11_+_129685707 | 0.04 |
ENST00000281441.3
|
TMEM45B
|
transmembrane protein 45B |
| chr21_-_46237883 | 0.04 |
ENST00000397893.3
|
SUMO3
|
small ubiquitin-like modifier 3 |
| chr15_-_77712429 | 0.04 |
ENST00000564328.1
ENST00000558305.1 |
PEAK1
|
pseudopodium-enriched atypical kinase 1 |
| chr13_+_31774073 | 0.04 |
ENST00000343307.4
|
B3GALTL
|
beta 1,3-galactosyltransferase-like |
| chr2_+_204193149 | 0.04 |
ENST00000422511.2
|
ABI2
|
abl-interactor 2 |
| chr11_-_30607819 | 0.04 |
ENST00000448418.2
|
MPPED2
|
metallophosphoesterase domain containing 2 |
| chr10_+_105881779 | 0.04 |
ENST00000369729.3
|
SFR1
|
SWI5-dependent recombination repair 1 |
| chr3_-_185826718 | 0.04 |
ENST00000413301.1
ENST00000421809.1 |
ETV5
|
ets variant 5 |
| chr3_-_133614421 | 0.04 |
ENST00000543906.1
|
RAB6B
|
RAB6B, member RAS oncogene family |
| chr18_-_5296001 | 0.04 |
ENST00000357006.4
|
ZBTB14
|
zinc finger and BTB domain containing 14 |
| chr11_-_76155618 | 0.04 |
ENST00000530759.1
|
RP11-111M22.3
|
RP11-111M22.3 |
| chr10_+_105881906 | 0.04 |
ENST00000369727.3
ENST00000336358.5 |
SFR1
|
SWI5-dependent recombination repair 1 |
| chr9_+_6758024 | 0.04 |
ENST00000442236.2
|
KDM4C
|
lysine (K)-specific demethylase 4C |
| chr6_+_135502501 | 0.04 |
ENST00000527615.1
ENST00000420123.2 ENST00000525369.1 ENST00000528774.1 ENST00000534121.1 ENST00000534044.1 ENST00000533624.1 |
MYB
|
v-myb avian myeloblastosis viral oncogene homolog |
| chr8_-_55014336 | 0.04 |
ENST00000343231.6
|
LYPLA1
|
lysophospholipase I |
| chr1_+_84543821 | 0.04 |
ENST00000370688.3
|
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr2_-_39664206 | 0.04 |
ENST00000484274.1
|
MAP4K3
|
mitogen-activated protein kinase kinase kinase kinase 3 |
| chr10_-_735553 | 0.04 |
ENST00000280886.6
ENST00000423550.1 |
DIP2C
|
DIP2 disco-interacting protein 2 homolog C (Drosophila) |
| chr6_-_18155285 | 0.04 |
ENST00000309983.4
|
TPMT
|
thiopurine S-methyltransferase |
| chr1_-_49242553 | 0.04 |
ENST00000371833.3
|
BEND5
|
BEN domain containing 5 |
| chr22_-_39548443 | 0.04 |
ENST00000401405.3
|
CBX7
|
chromobox homolog 7 |
| chr22_-_39096981 | 0.04 |
ENST00000427389.1
|
JOSD1
|
Josephin domain containing 1 |
| chr12_-_31479107 | 0.04 |
ENST00000542983.1
|
FAM60A
|
family with sequence similarity 60, member A |
| chr2_+_18059906 | 0.04 |
ENST00000304101.4
|
KCNS3
|
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3 |
| chr9_+_138371503 | 0.04 |
ENST00000604351.1
|
PPP1R26
|
protein phosphatase 1, regulatory subunit 26 |
| chr7_-_45960850 | 0.04 |
ENST00000381083.4
ENST00000381086.5 ENST00000275521.6 |
IGFBP3
|
insulin-like growth factor binding protein 3 |
| chr3_+_170075436 | 0.04 |
ENST00000476188.1
ENST00000259119.4 ENST00000426052.2 |
SKIL
|
SKI-like oncogene |
| chr18_-_45457478 | 0.04 |
ENST00000402690.2
ENST00000356825.4 |
SMAD2
|
SMAD family member 2 |
| chr13_-_36705425 | 0.04 |
ENST00000255448.4
ENST00000360631.3 ENST00000379892.4 |
DCLK1
|
doublecortin-like kinase 1 |
| chr1_-_42800860 | 0.04 |
ENST00000445886.1
ENST00000361346.1 ENST00000361776.1 |
FOXJ3
|
forkhead box J3 |
| chr8_-_23315131 | 0.04 |
ENST00000518718.1
|
ENTPD4
|
ectonucleoside triphosphate diphosphohydrolase 4 |
| chr2_+_64681103 | 0.04 |
ENST00000464281.1
|
LGALSL
|
lectin, galactoside-binding-like |
| chr17_-_66287310 | 0.04 |
ENST00000582867.1
|
SLC16A6
|
solute carrier family 16, member 6 |
| chr6_+_146056706 | 0.04 |
ENST00000603994.1
|
RP3-466P17.1
|
RP3-466P17.1 |
| chr7_-_27135591 | 0.04 |
ENST00000343060.4
ENST00000355633.5 |
HOXA1
|
homeobox A1 |
| chr10_+_65281123 | 0.04 |
ENST00000298249.4
ENST00000373758.4 |
REEP3
|
receptor accessory protein 3 |
| chr1_+_236305826 | 0.04 |
ENST00000366592.3
ENST00000366591.4 |
GPR137B
|
G protein-coupled receptor 137B |
| chr1_+_28099700 | 0.04 |
ENST00000440806.2
|
STX12
|
syntaxin 12 |
| chr1_+_6845497 | 0.04 |
ENST00000473578.1
ENST00000557126.1 |
CAMTA1
|
calmodulin binding transcription activator 1 |
| chr18_+_11103 | 0.04 |
ENST00000575820.1
ENST00000572573.1 |
AP005530.1
|
AP005530.1 |
| chr8_+_94712752 | 0.04 |
ENST00000522324.1
ENST00000522803.1 ENST00000423990.2 |
FAM92A1
|
family with sequence similarity 92, member A1 |
| chr1_+_182992545 | 0.04 |
ENST00000258341.4
|
LAMC1
|
laminin, gamma 1 (formerly LAMB2) |
| chr8_+_87354945 | 0.04 |
ENST00000517970.1
|
WWP1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chr10_-_104179682 | 0.04 |
ENST00000406432.1
|
PSD
|
pleckstrin and Sec7 domain containing |
| chr7_+_65338312 | 0.04 |
ENST00000434382.2
|
VKORC1L1
|
vitamin K epoxide reductase complex, subunit 1-like 1 |
| chr14_-_30396948 | 0.04 |
ENST00000331968.5
|
PRKD1
|
protein kinase D1 |
| chr6_-_93433538 | 0.04 |
ENST00000404689.2
|
RP11-127B16.1
|
RP11-127B16.1 |
| chr3_-_129612394 | 0.04 |
ENST00000505616.1
ENST00000426664.2 |
TMCC1
|
transmembrane and coiled-coil domain family 1 |
| chr5_-_178054014 | 0.04 |
ENST00000520957.1
|
CLK4
|
CDC-like kinase 4 |
| chr3_-_57113281 | 0.04 |
ENST00000468466.1
|
ARHGEF3
|
Rho guanine nucleotide exchange factor (GEF) 3 |
| chr5_+_61601965 | 0.04 |
ENST00000401507.3
|
KIF2A
|
kinesin heavy chain member 2A |
| chr17_+_35767301 | 0.04 |
ENST00000225396.6
ENST00000417170.1 ENST00000590005.1 ENST00000590957.1 |
TADA2A
|
transcriptional adaptor 2A |
| chr13_+_114238997 | 0.04 |
ENST00000538138.1
ENST00000375370.5 |
TFDP1
|
transcription factor Dp-1 |
| chr6_-_19804973 | 0.04 |
ENST00000457670.1
ENST00000607810.1 ENST00000606628.1 |
RP4-625H18.2
|
RP4-625H18.2 |
| chr4_+_1873155 | 0.04 |
ENST00000507820.1
ENST00000514045.1 |
WHSC1
|
Wolf-Hirschhorn syndrome candidate 1 |
| chr18_-_5296138 | 0.04 |
ENST00000400143.3
|
ZBTB14
|
zinc finger and BTB domain containing 14 |
| chr21_-_46238034 | 0.04 |
ENST00000332859.6
|
SUMO3
|
small ubiquitin-like modifier 3 |
| chr9_+_127020202 | 0.04 |
ENST00000373600.3
ENST00000320246.5 |
NEK6
|
NIMA-related kinase 6 |
| chr12_+_112451222 | 0.04 |
ENST00000552052.1
|
ERP29
|
endoplasmic reticulum protein 29 |
| chr9_-_8857776 | 0.04 |
ENST00000481079.1
|
PTPRD
|
protein tyrosine phosphatase, receptor type, D |
| chr5_+_134181755 | 0.04 |
ENST00000504727.1
ENST00000435259.2 ENST00000508791.1 |
C5orf24
|
chromosome 5 open reading frame 24 |
| chr4_+_1873100 | 0.04 |
ENST00000508803.1
|
WHSC1
|
Wolf-Hirschhorn syndrome candidate 1 |
| chr19_+_47104553 | 0.04 |
ENST00000598871.1
ENST00000594523.1 |
CALM3
|
calmodulin 3 (phosphorylase kinase, delta) |
| chr6_-_108582168 | 0.03 |
ENST00000426155.2
|
SNX3
|
sorting nexin 3 |
| chr2_+_121010324 | 0.03 |
ENST00000272519.5
|
RALB
|
v-ral simian leukemia viral oncogene homolog B |
| chr3_+_197477038 | 0.03 |
ENST00000426031.1
ENST00000424384.2 |
FYTTD1
|
forty-two-three domain containing 1 |
| chr8_-_90996459 | 0.03 |
ENST00000517337.1
ENST00000409330.1 |
NBN
|
nibrin |
| chrX_+_135579670 | 0.03 |
ENST00000218364.4
|
HTATSF1
|
HIV-1 Tat specific factor 1 |
| chr8_+_96146168 | 0.03 |
ENST00000519516.1
|
PLEKHF2
|
pleckstrin homology domain containing, family F (with FYVE domain) member 2 |
| chr1_-_236445251 | 0.03 |
ENST00000354619.5
ENST00000327333.8 |
ERO1LB
|
ERO1-like beta (S. cerevisiae) |
| chr5_+_121647877 | 0.03 |
ENST00000514497.2
ENST00000261367.7 |
SNCAIP
|
synuclein, alpha interacting protein |
| chr2_+_48010312 | 0.03 |
ENST00000540021.1
|
MSH6
|
mutS homolog 6 |
| chr3_-_49449521 | 0.03 |
ENST00000431929.1
ENST00000418115.1 |
RHOA
|
ras homolog family member A |
| chr6_+_96025341 | 0.03 |
ENST00000369293.1
ENST00000358812.4 |
MANEA
|
mannosidase, endo-alpha |
| chr12_+_32552451 | 0.03 |
ENST00000534526.2
|
FGD4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr2_+_121010370 | 0.03 |
ENST00000420510.1
|
RALB
|
v-ral simian leukemia viral oncogene homolog B |
| chr17_-_76837499 | 0.03 |
ENST00000592275.1
|
USP36
|
ubiquitin specific peptidase 36 |
| chr13_-_95953589 | 0.03 |
ENST00000538287.1
ENST00000376887.4 ENST00000412704.1 ENST00000536256.1 ENST00000431522.1 |
ABCC4
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 4 |
| chr8_-_144790224 | 0.03 |
ENST00000533508.1
ENST00000542437.1 |
CCDC166
|
coiled-coil domain containing 166 |
| chr1_+_94883991 | 0.03 |
ENST00000370214.4
|
ABCD3
|
ATP-binding cassette, sub-family D (ALD), member 3 |
| chr13_-_52026730 | 0.03 |
ENST00000420668.2
|
INTS6
|
integrator complex subunit 6 |
| chr21_+_47878757 | 0.03 |
ENST00000400274.1
ENST00000427143.2 ENST00000318711.7 ENST00000457905.3 ENST00000466639.1 ENST00000435722.3 ENST00000417564.2 |
DIP2A
|
DIP2 disco-interacting protein 2 homolog A (Drosophila) |
| chr5_+_142149932 | 0.03 |
ENST00000274498.4
|
ARHGAP26
|
Rho GTPase activating protein 26 |
| chr3_+_44596679 | 0.03 |
ENST00000426540.1
ENST00000431636.1 ENST00000341840.3 ENST00000273320.3 |
ZKSCAN7
|
zinc finger with KRAB and SCAN domains 7 |
| chr10_+_94608245 | 0.03 |
ENST00000443748.2
ENST00000260762.6 |
EXOC6
|
exocyst complex component 6 |
| chr1_+_185014647 | 0.03 |
ENST00000367509.4
|
RNF2
|
ring finger protein 2 |
| chr20_-_32700075 | 0.03 |
ENST00000374980.2
|
EIF2S2
|
eukaryotic translation initiation factor 2, subunit 2 beta, 38kDa |
| chr11_-_118023594 | 0.03 |
ENST00000529878.1
|
SCN4B
|
sodium channel, voltage-gated, type IV, beta subunit |
| chr7_+_77166592 | 0.03 |
ENST00000248594.6
|
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12 |
| chr8_-_42751728 | 0.03 |
ENST00000319104.3
ENST00000531440.1 |
RNF170
|
ring finger protein 170 |
| chr9_+_6757634 | 0.03 |
ENST00000543771.1
ENST00000401787.3 ENST00000381306.3 ENST00000381309.3 |
KDM4C
|
lysine (K)-specific demethylase 4C |
| chr20_+_45523227 | 0.03 |
ENST00000327619.5
ENST00000357410.3 |
EYA2
|
eyes absent homolog 2 (Drosophila) |
| chr5_+_170814803 | 0.03 |
ENST00000521672.1
ENST00000351986.6 ENST00000393820.2 ENST00000523622.1 |
NPM1
|
nucleophosmin (nucleolar phosphoprotein B23, numatrin) |
| chr4_-_78740769 | 0.03 |
ENST00000512485.1
|
CNOT6L
|
CCR4-NOT transcription complex, subunit 6-like |
| chr9_+_36572851 | 0.03 |
ENST00000298048.2
ENST00000538311.1 ENST00000536987.1 ENST00000545008.1 ENST00000536860.1 ENST00000536329.1 ENST00000541717.1 ENST00000543751.1 |
MELK
|
maternal embryonic leucine zipper kinase |
| chr3_-_138048682 | 0.03 |
ENST00000383180.2
|
NME9
|
NME/NM23 family member 9 |
| chr8_+_94712895 | 0.03 |
ENST00000523453.1
ENST00000520955.1 |
FAM92A1
|
family with sequence similarity 92, member A1 |
| chr6_-_86352982 | 0.03 |
ENST00000369622.3
|
SYNCRIP
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr1_+_174128536 | 0.03 |
ENST00000357444.6
ENST00000367689.3 |
RABGAP1L
|
RAB GTPase activating protein 1-like |
| chr4_-_2420335 | 0.03 |
ENST00000503000.1
|
ZFYVE28
|
zinc finger, FYVE domain containing 28 |
| chr13_+_111365602 | 0.03 |
ENST00000333219.7
|
ING1
|
inhibitor of growth family, member 1 |
| chr12_+_123717458 | 0.03 |
ENST00000253233.1
|
C12orf65
|
chromosome 12 open reading frame 65 |
| chrX_-_19905577 | 0.03 |
ENST00000379697.3
|
SH3KBP1
|
SH3-domain kinase binding protein 1 |
| chrX_-_16888276 | 0.03 |
ENST00000493145.1
|
RBBP7
|
retinoblastoma binding protein 7 |
| chr2_+_204193101 | 0.03 |
ENST00000430418.1
ENST00000424558.1 ENST00000261016.6 |
ABI2
|
abl-interactor 2 |
| chr1_+_178694300 | 0.03 |
ENST00000367635.3
|
RALGPS2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr14_+_89029366 | 0.03 |
ENST00000555799.1
ENST00000555755.1 ENST00000393514.5 |
ZC3H14
|
zinc finger CCCH-type containing 14 |
| chr3_-_118753566 | 0.03 |
ENST00000491903.1
|
IGSF11
|
immunoglobulin superfamily, member 11 |
| chr2_-_46385 | 0.03 |
ENST00000327669.4
|
FAM110C
|
family with sequence similarity 110, member C |
| chr1_-_29508321 | 0.03 |
ENST00000546138.1
|
SRSF4
|
serine/arginine-rich splicing factor 4 |
| chr17_-_4269768 | 0.03 |
ENST00000396981.2
|
UBE2G1
|
ubiquitin-conjugating enzyme E2G 1 |
| chr12_+_19282713 | 0.03 |
ENST00000299275.6
ENST00000539256.1 ENST00000538714.1 |
PLEKHA5
|
pleckstrin homology domain containing, family A member 5 |
| chr3_-_179169330 | 0.03 |
ENST00000232564.3
|
GNB4
|
guanine nucleotide binding protein (G protein), beta polypeptide 4 |
| chr10_+_21823243 | 0.03 |
ENST00000307729.7
ENST00000377091.2 |
MLLT10
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 10 |
| chr15_-_51914810 | 0.03 |
ENST00000543779.2
ENST00000449909.3 |
DMXL2
|
Dmx-like 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of ZBTB14
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 0.0 | 0.1 | GO:1902232 | regulation of positive thymic T cell selection(GO:1902232) |
| 0.0 | 0.1 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.1 | GO:0060178 | regulation of exocyst assembly(GO:0001928) regulation of exocyst localization(GO:0060178) |
| 0.0 | 0.1 | GO:0006679 | glucosylceramide biosynthetic process(GO:0006679) |
| 0.0 | 0.1 | GO:0021861 | forebrain radial glial cell differentiation(GO:0021861) |
| 0.0 | 0.1 | GO:0044830 | modulation by host of viral RNA genome replication(GO:0044830) positive regulation of intracellular transport of viral material(GO:1901254) |
| 0.0 | 0.1 | GO:1900110 | negative regulation of histone H3-K9 dimethylation(GO:1900110) |
| 0.0 | 0.2 | GO:0032485 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.1 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 | 0.1 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.0 | 0.1 | GO:0071140 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 0.0 | 0.2 | GO:0000414 | regulation of histone H3-K36 methylation(GO:0000414) |
| 0.0 | 0.1 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.0 | 0.1 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.1 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.0 | 0.0 | GO:1904897 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.0 | 0.0 | GO:2000820 | pathogenesis(GO:0009405) negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.0 | 0.1 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.1 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.0 | 0.1 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.1 | GO:0021644 | vagus nerve morphogenesis(GO:0021644) |
| 0.0 | 0.1 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.0 | 0.0 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.0 | 0.1 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.0 | GO:0070676 | intralumenal vesicle formation(GO:0070676) |
| 0.0 | 0.1 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.0 | GO:0035750 | protein localization to myelin sheath abaxonal region(GO:0035750) |
| 0.0 | 0.1 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.0 | 0.1 | GO:1901727 | positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway(GO:0038033) positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 0.0 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.0 | 0.1 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.0 | GO:0007387 | anterior compartment pattern formation(GO:0007387) posterior compartment specification(GO:0007388) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1902737 | dendritic filopodium(GO:1902737) |
| 0.0 | 0.1 | GO:0032798 | Swi5-Sfr1 complex(GO:0032798) |
| 0.0 | 0.0 | GO:0097637 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.0 | 0.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.1 | GO:0097362 | MCM8-MCM9 complex(GO:0097362) |
| 0.0 | 0.1 | GO:0043291 | RAVE complex(GO:0043291) |
| 0.0 | 0.0 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.1 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.1 | GO:0032301 | MutSalpha complex(GO:0032301) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.2 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.0 | 0.1 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 0.1 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.0 | 0.1 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.1 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.2 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.0 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.0 | 0.1 | GO:0032143 | single thymine insertion binding(GO:0032143) |
| 0.0 | 0.2 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.0 | GO:0047322 | [hydroxymethylglutaryl-CoA reductase (NADPH)] kinase activity(GO:0047322) [acetyl-CoA carboxylase] kinase activity(GO:0050405) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |