Project
A549 cells infected with IAV Analysis Results (GEO series: GSE147507)
Navigation
Downloads
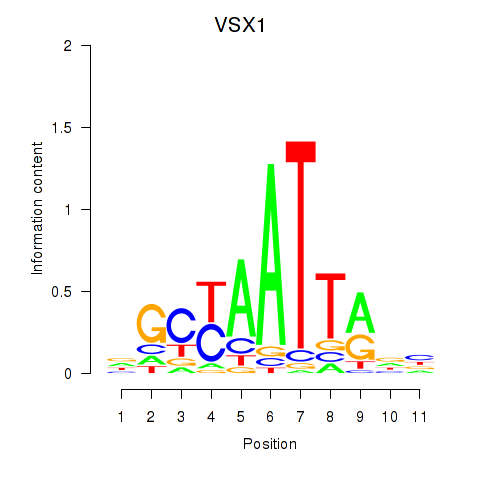
Results for VSX1
Z-value: 0.52
Transcription factors associated with VSX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
VSX1
|
ENSG00000100987.10 | visual system homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| VSX1 | hg19_v2_chr20_-_25062767_25062779 | 0.88 | 1.2e-01 | Click! |
Activity profile of VSX1 motif
Sorted Z-values of VSX1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_-_47509994 | 0.69 |
ENST00000343894.4
|
ELK1
|
ELK1, member of ETS oncogene family |
| chrX_-_47509887 | 0.38 |
ENST00000247161.3
ENST00000592066.1 ENST00000376983.3 |
ELK1
|
ELK1, member of ETS oncogene family |
| chr11_+_43702322 | 0.31 |
ENST00000395700.4
|
HSD17B12
|
hydroxysteroid (17-beta) dehydrogenase 12 |
| chr4_+_147096837 | 0.28 |
ENST00000296581.5
ENST00000502781.1 |
LSM6
|
LSM6 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr13_+_110958124 | 0.26 |
ENST00000400163.2
|
COL4A2
|
collagen, type IV, alpha 2 |
| chr3_-_120400960 | 0.26 |
ENST00000476082.2
|
HGD
|
homogentisate 1,2-dioxygenase |
| chr7_-_86849836 | 0.25 |
ENST00000455575.1
|
TMEM243
|
transmembrane protein 243, mitochondrial |
| chr19_-_54618650 | 0.24 |
ENST00000391757.1
|
TFPT
|
TCF3 (E2A) fusion partner (in childhood Leukemia) |
| chr17_-_7307358 | 0.22 |
ENST00000576017.1
ENST00000302422.3 ENST00000535512.1 |
TMEM256
TMEM256-PLSCR3
|
transmembrane protein 256 TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr2_+_240323439 | 0.21 |
ENST00000428471.1
ENST00000413029.1 |
AC062017.1
|
Uncharacterized protein |
| chr19_-_40324767 | 0.21 |
ENST00000601972.1
ENST00000430012.2 ENST00000323039.5 ENST00000348817.3 |
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr16_+_68279207 | 0.20 |
ENST00000413021.2
ENST00000565744.1 ENST00000219345.5 |
PLA2G15
|
phospholipase A2, group XV |
| chr9_+_131799213 | 0.20 |
ENST00000358369.4
ENST00000406926.2 ENST00000277475.5 ENST00000450073.1 |
FAM73B
|
family with sequence similarity 73, member B |
| chrX_-_77150911 | 0.19 |
ENST00000373336.3
|
MAGT1
|
magnesium transporter 1 |
| chr19_+_9945962 | 0.19 |
ENST00000587625.1
ENST00000247970.4 ENST00000588695.1 |
PIN1
|
peptidylprolyl cis/trans isomerase, NIMA-interacting 1 |
| chr14_+_23790655 | 0.19 |
ENST00000397276.2
|
PABPN1
|
poly(A) binding protein, nuclear 1 |
| chr7_+_99699179 | 0.19 |
ENST00000438383.1
ENST00000429084.1 ENST00000359593.4 ENST00000439416.1 |
AP4M1
|
adaptor-related protein complex 4, mu 1 subunit |
| chr2_-_131099897 | 0.19 |
ENST00000409127.1
ENST00000437688.2 ENST00000259229.2 |
CCDC115
|
coiled-coil domain containing 115 |
| chr13_-_110438914 | 0.18 |
ENST00000375856.3
|
IRS2
|
insulin receptor substrate 2 |
| chr19_+_50270219 | 0.18 |
ENST00000354293.5
ENST00000359032.5 |
AP2A1
|
adaptor-related protein complex 2, alpha 1 subunit |
| chr19_+_39421556 | 0.18 |
ENST00000407800.2
ENST00000402029.3 |
MRPS12
|
mitochondrial ribosomal protein S12 |
| chr11_-_66056596 | 0.18 |
ENST00000471387.2
ENST00000359461.6 ENST00000376901.4 |
YIF1A
|
Yip1 interacting factor homolog A (S. cerevisiae) |
| chr6_-_32145861 | 0.17 |
ENST00000336984.6
|
AGPAT1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr16_+_68279256 | 0.17 |
ENST00000564827.2
ENST00000566188.1 ENST00000444212.2 ENST00000568082.1 |
PLA2G15
|
phospholipase A2, group XV |
| chr1_+_159750776 | 0.16 |
ENST00000368107.1
|
DUSP23
|
dual specificity phosphatase 23 |
| chr2_-_73460334 | 0.16 |
ENST00000258083.2
|
PRADC1
|
protease-associated domain containing 1 |
| chr15_+_89631647 | 0.16 |
ENST00000569550.1
ENST00000565066.1 ENST00000565973.1 |
ABHD2
|
abhydrolase domain containing 2 |
| chr12_+_110011571 | 0.16 |
ENST00000539696.1
ENST00000228510.3 ENST00000392727.3 |
MVK
|
mevalonate kinase |
| chr1_-_167905225 | 0.16 |
ENST00000367846.4
|
MPC2
|
mitochondrial pyruvate carrier 2 |
| chr17_-_40829026 | 0.16 |
ENST00000412503.1
|
PLEKHH3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr17_+_62223320 | 0.16 |
ENST00000580828.1
ENST00000582965.1 |
SNORA76
|
small nucleolar RNA, H/ACA box 76 |
| chr1_-_154946792 | 0.16 |
ENST00000412170.1
|
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr3_-_42003613 | 0.15 |
ENST00000414606.1
|
ULK4
|
unc-51 like kinase 4 |
| chr17_-_40828969 | 0.15 |
ENST00000591022.1
ENST00000587627.1 ENST00000293349.6 |
PLEKHH3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr7_+_99699280 | 0.15 |
ENST00000421755.1
|
AP4M1
|
adaptor-related protein complex 4, mu 1 subunit |
| chr6_-_30685214 | 0.15 |
ENST00000425072.1
|
MDC1
|
mediator of DNA-damage checkpoint 1 |
| chr2_-_70520539 | 0.14 |
ENST00000482975.2
ENST00000438261.1 |
SNRPG
|
small nuclear ribonucleoprotein polypeptide G |
| chr2_+_74881432 | 0.14 |
ENST00000453930.1
|
SEMA4F
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4F |
| chr19_+_11485333 | 0.14 |
ENST00000312423.2
|
SWSAP1
|
SWIM-type zinc finger 7 associated protein 1 |
| chr2_-_98280383 | 0.14 |
ENST00000289228.5
|
ACTR1B
|
ARP1 actin-related protein 1 homolog B, centractin beta (yeast) |
| chr19_+_38893751 | 0.14 |
ENST00000588262.1
ENST00000252530.5 ENST00000343358.7 |
FAM98C
|
family with sequence similarity 98, member C |
| chr6_+_27861190 | 0.14 |
ENST00000303806.4
|
HIST1H2BO
|
histone cluster 1, H2bo |
| chr18_+_3411595 | 0.14 |
ENST00000552383.1
|
TGIF1
|
TGFB-induced factor homeobox 1 |
| chr11_-_133826852 | 0.14 |
ENST00000533871.2
ENST00000321016.8 |
IGSF9B
|
immunoglobulin superfamily, member 9B |
| chr16_-_2264779 | 0.13 |
ENST00000333503.7
|
PGP
|
phosphoglycolate phosphatase |
| chr7_+_100136811 | 0.13 |
ENST00000300176.4
ENST00000262935.4 |
AGFG2
|
ArfGAP with FG repeats 2 |
| chr3_-_186288097 | 0.13 |
ENST00000446782.1
|
TBCCD1
|
TBCC domain containing 1 |
| chr7_-_77427676 | 0.13 |
ENST00000257663.3
|
TMEM60
|
transmembrane protein 60 |
| chr19_-_54663473 | 0.13 |
ENST00000222224.3
|
LENG1
|
leukocyte receptor cluster (LRC) member 1 |
| chr19_-_15236173 | 0.13 |
ENST00000527093.1
|
ILVBL
|
ilvB (bacterial acetolactate synthase)-like |
| chr17_-_72869086 | 0.13 |
ENST00000581530.1
ENST00000420580.2 ENST00000455107.2 ENST00000413947.2 ENST00000581219.1 ENST00000582944.1 |
FDXR
|
ferredoxin reductase |
| chr19_-_39340563 | 0.13 |
ENST00000601813.1
|
HNRNPL
|
heterogeneous nuclear ribonucleoprotein L |
| chr4_+_96012585 | 0.13 |
ENST00000502683.1
|
BMPR1B
|
bone morphogenetic protein receptor, type IB |
| chr17_+_46970134 | 0.12 |
ENST00000503641.1
ENST00000514808.1 |
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr17_-_72869140 | 0.12 |
ENST00000583917.1
ENST00000293195.5 ENST00000442102.2 |
FDXR
|
ferredoxin reductase |
| chr19_-_39421377 | 0.12 |
ENST00000430193.3
ENST00000600042.1 ENST00000221431.6 |
SARS2
|
seryl-tRNA synthetase 2, mitochondrial |
| chr16_+_69345243 | 0.12 |
ENST00000254950.11
|
VPS4A
|
vacuolar protein sorting 4 homolog A (S. cerevisiae) |
| chr16_-_67217844 | 0.12 |
ENST00000563902.1
ENST00000561621.1 ENST00000290881.7 |
KIAA0895L
|
KIAA0895-like |
| chr16_-_30773372 | 0.12 |
ENST00000545825.1
ENST00000541260.1 |
C16orf93
|
chromosome 16 open reading frame 93 |
| chr14_-_24711764 | 0.12 |
ENST00000557921.1
ENST00000558476.1 |
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr19_-_40324255 | 0.12 |
ENST00000593685.1
ENST00000600611.1 |
DYRK1B
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr19_+_46498704 | 0.12 |
ENST00000595358.1
ENST00000594672.1 ENST00000536603.1 |
CCDC61
|
coiled-coil domain containing 61 |
| chr17_-_8059638 | 0.12 |
ENST00000584202.1
ENST00000354903.5 ENST00000577253.1 |
PER1
|
period circadian clock 1 |
| chr5_+_148737562 | 0.12 |
ENST00000274569.4
|
PCYOX1L
|
prenylcysteine oxidase 1 like |
| chr22_-_24316648 | 0.11 |
ENST00000403754.3
ENST00000430101.2 ENST00000398344.4 |
DDT
|
D-dopachrome tautomerase |
| chr5_-_176738883 | 0.11 |
ENST00000513169.1
ENST00000423571.2 ENST00000502529.1 ENST00000427908.2 |
MXD3
|
MAX dimerization protein 3 |
| chr19_+_56652643 | 0.11 |
ENST00000586123.1
|
ZNF444
|
zinc finger protein 444 |
| chr3_+_149191723 | 0.11 |
ENST00000305354.4
|
TM4SF4
|
transmembrane 4 L six family member 4 |
| chr3_-_61237050 | 0.11 |
ENST00000476844.1
ENST00000488467.1 ENST00000492590.1 ENST00000468189.1 |
FHIT
|
fragile histidine triad |
| chr13_+_19756173 | 0.10 |
ENST00000382988.2
|
RP11-408E5.4
|
RP11-408E5.4 |
| chr22_-_38240316 | 0.10 |
ENST00000411961.2
ENST00000434930.1 |
ANKRD54
|
ankyrin repeat domain 54 |
| chr15_+_89631381 | 0.10 |
ENST00000352732.5
|
ABHD2
|
abhydrolase domain containing 2 |
| chr11_+_67798363 | 0.10 |
ENST00000525628.1
|
NDUFS8
|
NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase) |
| chr4_+_169633310 | 0.10 |
ENST00000510998.1
|
PALLD
|
palladin, cytoskeletal associated protein |
| chr8_+_145159415 | 0.10 |
ENST00000534585.1
|
MAF1
|
MAF1 homolog (S. cerevisiae) |
| chr14_-_104181771 | 0.10 |
ENST00000554913.1
ENST00000554974.1 ENST00000553361.1 ENST00000555055.1 ENST00000555964.1 ENST00000556682.1 ENST00000445556.1 ENST00000553332.1 ENST00000352127.7 |
XRCC3
|
X-ray repair complementing defective repair in Chinese hamster cells 3 |
| chr6_-_26043885 | 0.10 |
ENST00000357905.2
|
HIST1H2BB
|
histone cluster 1, H2bb |
| chr22_-_30960876 | 0.10 |
ENST00000401975.1
ENST00000428682.1 ENST00000423299.1 |
GAL3ST1
|
galactose-3-O-sulfotransferase 1 |
| chr19_-_14606900 | 0.10 |
ENST00000393029.3
ENST00000393028.1 ENST00000393033.4 ENST00000345425.2 ENST00000586027.1 ENST00000591349.1 ENST00000587210.1 |
GIPC1
|
GIPC PDZ domain containing family, member 1 |
| chr8_+_145159376 | 0.10 |
ENST00000322428.5
|
MAF1
|
MAF1 homolog (S. cerevisiae) |
| chr2_-_70780770 | 0.10 |
ENST00000444975.1
ENST00000445399.1 ENST00000418333.2 |
TGFA
|
transforming growth factor, alpha |
| chr7_-_111424506 | 0.09 |
ENST00000450156.1
ENST00000494651.2 |
DOCK4
|
dedicator of cytokinesis 4 |
| chr14_+_104182061 | 0.09 |
ENST00000216602.6
|
ZFYVE21
|
zinc finger, FYVE domain containing 21 |
| chr10_-_5046042 | 0.09 |
ENST00000421196.3
ENST00000455190.1 |
AKR1C2
|
aldo-keto reductase family 1, member C2 |
| chr17_+_46970127 | 0.09 |
ENST00000355938.5
|
ATP5G1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr11_+_560956 | 0.09 |
ENST00000397582.3
ENST00000344375.4 ENST00000397583.3 |
RASSF7
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7 |
| chr6_-_27860956 | 0.09 |
ENST00000359611.2
|
HIST1H2AM
|
histone cluster 1, H2am |
| chrM_+_10464 | 0.09 |
ENST00000361335.1
|
MT-ND4L
|
mitochondrially encoded NADH dehydrogenase 4L |
| chr16_-_11036300 | 0.09 |
ENST00000331808.4
|
DEXI
|
Dexi homolog (mouse) |
| chr7_+_916183 | 0.09 |
ENST00000265857.3
|
GET4
|
golgi to ER traffic protein 4 homolog (S. cerevisiae) |
| chr17_-_1532106 | 0.09 |
ENST00000301335.5
ENST00000382147.4 |
SLC43A2
|
solute carrier family 43 (amino acid system L transporter), member 2 |
| chr19_-_47616992 | 0.09 |
ENST00000253048.5
|
ZC3H4
|
zinc finger CCCH-type containing 4 |
| chr19_-_49622348 | 0.09 |
ENST00000408991.2
|
C19orf73
|
chromosome 19 open reading frame 73 |
| chr1_-_154946825 | 0.09 |
ENST00000368453.4
ENST00000368450.1 ENST00000366442.2 |
SHC1
|
SHC (Src homology 2 domain containing) transforming protein 1 |
| chr12_-_56694083 | 0.09 |
ENST00000552688.1
ENST00000548041.1 ENST00000551137.1 ENST00000551968.1 ENST00000542324.2 ENST00000546930.1 ENST00000549221.1 ENST00000550159.1 ENST00000550734.1 |
CS
|
citrate synthase |
| chr1_-_36916066 | 0.08 |
ENST00000315643.9
|
OSCP1
|
organic solute carrier partner 1 |
| chr1_-_21625486 | 0.08 |
ENST00000481130.2
|
ECE1
|
endothelin converting enzyme 1 |
| chr19_-_15236470 | 0.08 |
ENST00000533747.1
ENST00000598709.1 ENST00000534378.1 |
ILVBL
|
ilvB (bacterial acetolactate synthase)-like |
| chr11_+_119205222 | 0.08 |
ENST00000311413.4
|
RNF26
|
ring finger protein 26 |
| chr5_+_159343688 | 0.08 |
ENST00000306675.3
|
ADRA1B
|
adrenoceptor alpha 1B |
| chr9_-_140095186 | 0.08 |
ENST00000409012.4
|
TPRN
|
taperin |
| chr2_-_240322643 | 0.08 |
ENST00000345617.3
|
HDAC4
|
histone deacetylase 4 |
| chr13_-_19755975 | 0.08 |
ENST00000400113.3
|
TUBA3C
|
tubulin, alpha 3c |
| chr3_-_120461353 | 0.07 |
ENST00000483733.1
|
RABL3
|
RAB, member of RAS oncogene family-like 3 |
| chr16_+_30773610 | 0.07 |
ENST00000566811.1
|
RNF40
|
ring finger protein 40, E3 ubiquitin protein ligase |
| chr22_-_38240412 | 0.07 |
ENST00000215941.4
|
ANKRD54
|
ankyrin repeat domain 54 |
| chr1_-_78444738 | 0.07 |
ENST00000436586.2
ENST00000370768.2 |
FUBP1
|
far upstream element (FUSE) binding protein 1 |
| chr16_+_53133070 | 0.07 |
ENST00000565832.1
|
CHD9
|
chromodomain helicase DNA binding protein 9 |
| chr19_+_41770236 | 0.07 |
ENST00000392006.3
|
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr17_-_47841485 | 0.07 |
ENST00000506156.1
ENST00000240364.2 |
FAM117A
|
family with sequence similarity 117, member A |
| chr11_-_33913708 | 0.07 |
ENST00000257818.2
|
LMO2
|
LIM domain only 2 (rhombotin-like 1) |
| chr11_-_66056478 | 0.07 |
ENST00000431556.2
ENST00000528575.1 |
YIF1A
|
Yip1 interacting factor homolog A (S. cerevisiae) |
| chr19_+_56652686 | 0.07 |
ENST00000592949.1
|
ZNF444
|
zinc finger protein 444 |
| chr19_+_7895074 | 0.07 |
ENST00000270530.4
|
EVI5L
|
ecotropic viral integration site 5-like |
| chr1_+_6052700 | 0.07 |
ENST00000378092.1
ENST00000445501.1 |
KCNAB2
|
potassium voltage-gated channel, shaker-related subfamily, beta member 2 |
| chr7_-_111424462 | 0.07 |
ENST00000437129.1
|
DOCK4
|
dedicator of cytokinesis 4 |
| chr9_-_116840728 | 0.07 |
ENST00000265132.3
|
AMBP
|
alpha-1-microglobulin/bikunin precursor |
| chr7_-_148725544 | 0.07 |
ENST00000413966.1
|
PDIA4
|
protein disulfide isomerase family A, member 4 |
| chr16_+_22501658 | 0.07 |
ENST00000415833.2
|
NPIPB5
|
nuclear pore complex interacting protein family, member B5 |
| chr1_-_78444776 | 0.07 |
ENST00000370767.1
ENST00000421641.1 |
FUBP1
|
far upstream element (FUSE) binding protein 1 |
| chr19_+_41770349 | 0.07 |
ENST00000602130.1
|
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr20_-_32274179 | 0.07 |
ENST00000343380.5
|
E2F1
|
E2F transcription factor 1 |
| chr14_-_24711865 | 0.07 |
ENST00000399423.4
ENST00000267415.7 |
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr8_-_145159083 | 0.07 |
ENST00000398712.2
|
SHARPIN
|
SHANK-associated RH domain interactor |
| chr17_+_8191815 | 0.07 |
ENST00000226105.6
ENST00000407006.4 ENST00000580434.1 ENST00000439238.3 |
RANGRF
|
RAN guanine nucleotide release factor |
| chr19_+_45394477 | 0.06 |
ENST00000252487.5
ENST00000405636.2 ENST00000592434.1 ENST00000426677.2 ENST00000589649.1 |
TOMM40
|
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chr1_+_84630574 | 0.06 |
ENST00000413538.1
ENST00000417530.1 |
PRKACB
|
protein kinase, cAMP-dependent, catalytic, beta |
| chr3_-_44519131 | 0.06 |
ENST00000425708.2
ENST00000396077.2 |
ZNF445
|
zinc finger protein 445 |
| chr12_+_7014126 | 0.06 |
ENST00000415834.1
ENST00000436789.1 |
LRRC23
|
leucine rich repeat containing 23 |
| chr8_+_97773202 | 0.06 |
ENST00000519484.1
|
CPQ
|
carboxypeptidase Q |
| chr2_+_219135115 | 0.06 |
ENST00000248451.3
ENST00000273077.4 |
PNKD
|
paroxysmal nonkinesigenic dyskinesia |
| chr10_+_99205959 | 0.06 |
ENST00000352634.4
ENST00000353979.3 ENST00000370842.2 ENST00000345745.5 |
ZDHHC16
|
zinc finger, DHHC-type containing 16 |
| chr10_+_5005598 | 0.06 |
ENST00000442997.1
|
AKR1C1
|
aldo-keto reductase family 1, member C1 |
| chr5_-_137374288 | 0.06 |
ENST00000514310.1
|
FAM13B
|
family with sequence similarity 13, member B |
| chr19_+_4007644 | 0.06 |
ENST00000262971.2
|
PIAS4
|
protein inhibitor of activated STAT, 4 |
| chr15_-_53002007 | 0.06 |
ENST00000561490.1
|
FAM214A
|
family with sequence similarity 214, member A |
| chr7_+_99717230 | 0.06 |
ENST00000262932.3
|
CNPY4
|
canopy FGF signaling regulator 4 |
| chr5_-_13944652 | 0.06 |
ENST00000265104.4
|
DNAH5
|
dynein, axonemal, heavy chain 5 |
| chrX_-_153363188 | 0.06 |
ENST00000303391.6
|
MECP2
|
methyl CpG binding protein 2 (Rett syndrome) |
| chr6_+_30687978 | 0.06 |
ENST00000327892.8
ENST00000435534.1 |
TUBB
|
tubulin, beta class I |
| chr19_+_34287751 | 0.06 |
ENST00000590771.1
ENST00000589786.1 ENST00000284006.6 ENST00000588881.1 |
KCTD15
|
potassium channel tetramerization domain containing 15 |
| chr9_+_140135665 | 0.06 |
ENST00000340384.4
|
TUBB4B
|
tubulin, beta 4B class IVb |
| chr14_-_24711806 | 0.06 |
ENST00000540705.1
ENST00000538777.1 ENST00000558566.1 ENST00000559019.1 |
TINF2
|
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr16_+_3184924 | 0.05 |
ENST00000574902.1
ENST00000396878.3 |
ZNF213
|
zinc finger protein 213 |
| chr17_+_74722912 | 0.05 |
ENST00000589977.1
ENST00000591571.1 ENST00000592849.1 ENST00000586738.1 ENST00000588783.1 ENST00000588563.1 ENST00000586752.1 ENST00000588302.1 ENST00000590964.1 ENST00000341249.6 ENST00000588822.1 |
METTL23
|
methyltransferase like 23 |
| chr17_-_79650818 | 0.05 |
ENST00000397498.4
|
ARL16
|
ADP-ribosylation factor-like 16 |
| chr10_+_99205894 | 0.05 |
ENST00000370854.3
ENST00000393760.1 ENST00000414567.1 ENST00000370846.4 |
ZDHHC16
|
zinc finger, DHHC-type containing 16 |
| chr19_-_54619006 | 0.05 |
ENST00000391759.1
|
TFPT
|
TCF3 (E2A) fusion partner (in childhood Leukemia) |
| chr6_+_26124373 | 0.05 |
ENST00000377791.2
ENST00000602637.1 |
HIST1H2AC
|
histone cluster 1, H2ac |
| chr6_-_62996066 | 0.05 |
ENST00000281156.4
|
KHDRBS2
|
KH domain containing, RNA binding, signal transduction associated 2 |
| chr1_-_20987851 | 0.05 |
ENST00000464364.1
ENST00000602624.2 |
DDOST
|
dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit (non-catalytic) |
| chr8_-_29120580 | 0.05 |
ENST00000524189.1
|
KIF13B
|
kinesin family member 13B |
| chr16_-_87350970 | 0.05 |
ENST00000567970.1
|
C16orf95
|
chromosome 16 open reading frame 95 |
| chr14_-_71276211 | 0.05 |
ENST00000381250.4
ENST00000555993.2 |
MAP3K9
|
mitogen-activated protein kinase kinase kinase 9 |
| chr19_-_2256405 | 0.05 |
ENST00000300961.6
|
JSRP1
|
junctional sarcoplasmic reticulum protein 1 |
| chr10_+_98064085 | 0.05 |
ENST00000419175.1
ENST00000371174.2 |
DNTT
|
DNA nucleotidylexotransferase |
| chr4_-_175204765 | 0.05 |
ENST00000513696.1
ENST00000503293.1 |
FBXO8
|
F-box protein 8 |
| chr15_-_64673665 | 0.05 |
ENST00000300035.4
|
KIAA0101
|
KIAA0101 |
| chr6_+_42018614 | 0.05 |
ENST00000465926.1
ENST00000482432.1 |
TAF8
|
TAF8 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 43kDa |
| chr12_-_57039739 | 0.05 |
ENST00000552959.1
ENST00000551020.1 ENST00000553007.2 ENST00000552919.1 ENST00000552104.1 ENST00000262030.3 |
ATP5B
|
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide |
| chr6_-_26199471 | 0.05 |
ENST00000341023.1
|
HIST1H2AD
|
histone cluster 1, H2ad |
| chr15_-_64673630 | 0.05 |
ENST00000558008.1
ENST00000559519.1 ENST00000380258.2 |
KIAA0101
|
KIAA0101 |
| chr6_+_36164487 | 0.05 |
ENST00000357641.6
|
BRPF3
|
bromodomain and PHD finger containing, 3 |
| chr15_-_37393406 | 0.05 |
ENST00000338564.5
ENST00000558313.1 ENST00000340545.5 |
MEIS2
|
Meis homeobox 2 |
| chr11_+_3819049 | 0.05 |
ENST00000396986.2
ENST00000300730.6 ENST00000532535.1 ENST00000396993.4 ENST00000396991.2 ENST00000532523.1 ENST00000459679.1 ENST00000464261.1 ENST00000464906.2 ENST00000464441.1 |
PGAP2
|
post-GPI attachment to proteins 2 |
| chr6_-_32157947 | 0.05 |
ENST00000375050.4
|
PBX2
|
pre-B-cell leukemia homeobox 2 |
| chr19_-_14247365 | 0.05 |
ENST00000592798.1
ENST00000474890.1 |
ASF1B
|
anti-silencing function 1B histone chaperone |
| chr19_+_39881951 | 0.04 |
ENST00000315588.5
ENST00000594368.1 ENST00000599213.2 ENST00000596297.1 |
MED29
|
mediator complex subunit 29 |
| chr7_-_99716952 | 0.04 |
ENST00000523306.1
ENST00000344095.4 ENST00000417349.1 ENST00000493322.1 ENST00000520135.1 ENST00000418432.2 ENST00000460673.2 ENST00000452041.1 ENST00000452438.2 ENST00000451699.1 ENST00000453269.2 |
TAF6
|
TAF6 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 80kDa |
| chr16_-_4784128 | 0.04 |
ENST00000592711.1
ENST00000590147.1 ENST00000304283.4 ENST00000592190.1 ENST00000589065.1 ENST00000585773.1 ENST00000450067.2 ENST00000592698.1 ENST00000586166.1 ENST00000586605.1 ENST00000592421.1 |
ANKS3
|
ankyrin repeat and sterile alpha motif domain containing 3 |
| chr19_-_11266437 | 0.04 |
ENST00000586708.1
ENST00000591396.1 ENST00000592967.1 ENST00000585486.1 ENST00000585567.1 |
SPC24
|
SPC24, NDC80 kinetochore complex component |
| chr7_-_137686791 | 0.04 |
ENST00000452463.1
ENST00000330387.6 ENST00000456390.1 |
CREB3L2
|
cAMP responsive element binding protein 3-like 2 |
| chr2_-_71454185 | 0.04 |
ENST00000244221.8
|
PAIP2B
|
poly(A) binding protein interacting protein 2B |
| chr14_+_32798462 | 0.04 |
ENST00000280979.4
|
AKAP6
|
A kinase (PRKA) anchor protein 6 |
| chr13_-_34250861 | 0.04 |
ENST00000445227.1
ENST00000454681.2 |
RP11-141M1.3
|
RP11-141M1.3 |
| chr2_-_240322685 | 0.04 |
ENST00000544989.1
|
HDAC4
|
histone deacetylase 4 |
| chr16_+_23652700 | 0.04 |
ENST00000300087.2
|
DCTN5
|
dynactin 5 (p25) |
| chr1_-_112281875 | 0.04 |
ENST00000527621.1
ENST00000534365.1 ENST00000357260.5 |
FAM212B
|
family with sequence similarity 212, member B |
| chr6_-_18264406 | 0.04 |
ENST00000515742.1
|
DEK
|
DEK oncogene |
| chrX_+_37639302 | 0.04 |
ENST00000545017.1
ENST00000536160.1 |
CYBB
|
cytochrome b-245, beta polypeptide |
| chr3_-_42003479 | 0.04 |
ENST00000420927.1
|
ULK4
|
unc-51 like kinase 4 |
| chr17_-_74163159 | 0.04 |
ENST00000591615.1
|
RNF157
|
ring finger protein 157 |
| chr11_-_62521614 | 0.04 |
ENST00000527994.1
ENST00000394807.3 |
ZBTB3
|
zinc finger and BTB domain containing 3 |
| chr4_-_109541539 | 0.04 |
ENST00000509984.1
ENST00000507248.1 ENST00000506795.1 |
RPL34-AS1
|
RPL34 antisense RNA 1 (head to head) |
| chr5_+_31193847 | 0.04 |
ENST00000514738.1
ENST00000265071.2 |
CDH6
|
cadherin 6, type 2, K-cadherin (fetal kidney) |
| chr15_+_83478370 | 0.04 |
ENST00000286760.4
|
WHAMM
|
WAS protein homolog associated with actin, golgi membranes and microtubules |
| chr9_+_34646624 | 0.04 |
ENST00000450095.2
ENST00000556278.1 |
GALT
GALT
|
galactose-1-phosphate uridylyltransferase Uncharacterized protein |
| chr4_-_143226979 | 0.04 |
ENST00000514525.1
|
INPP4B
|
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr19_+_41770269 | 0.04 |
ENST00000378215.4
|
HNRNPUL1
|
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr6_+_33422343 | 0.04 |
ENST00000395064.2
|
ZBTB9
|
zinc finger and BTB domain containing 9 |
| chr12_-_110011288 | 0.04 |
ENST00000540016.1
ENST00000266839.5 |
MMAB
|
methylmalonic aciduria (cobalamin deficiency) cblB type |
| chr13_-_30160925 | 0.04 |
ENST00000450494.1
|
SLC7A1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr5_+_98109322 | 0.04 |
ENST00000513185.1
|
RGMB
|
repulsive guidance molecule family member b |
| chr16_-_31085514 | 0.04 |
ENST00000300849.4
|
ZNF668
|
zinc finger protein 668 |
| chrX_+_37639264 | 0.04 |
ENST00000378588.4
|
CYBB
|
cytochrome b-245, beta polypeptide |
| chr12_-_6961050 | 0.04 |
ENST00000538862.2
|
CDCA3
|
cell division cycle associated 3 |
| chr12_+_118573663 | 0.03 |
ENST00000261313.2
|
PEBP1
|
phosphatidylethanolamine binding protein 1 |
| chr1_+_206138457 | 0.03 |
ENST00000367128.3
ENST00000431655.2 |
FAM72A
|
family with sequence similarity 72, member A |
| chr4_-_119759795 | 0.03 |
ENST00000419654.2
|
SEC24D
|
SEC24 family member D |
| chr19_-_11266471 | 0.03 |
ENST00000592540.1
|
SPC24
|
SPC24, NDC80 kinetochore complex component |
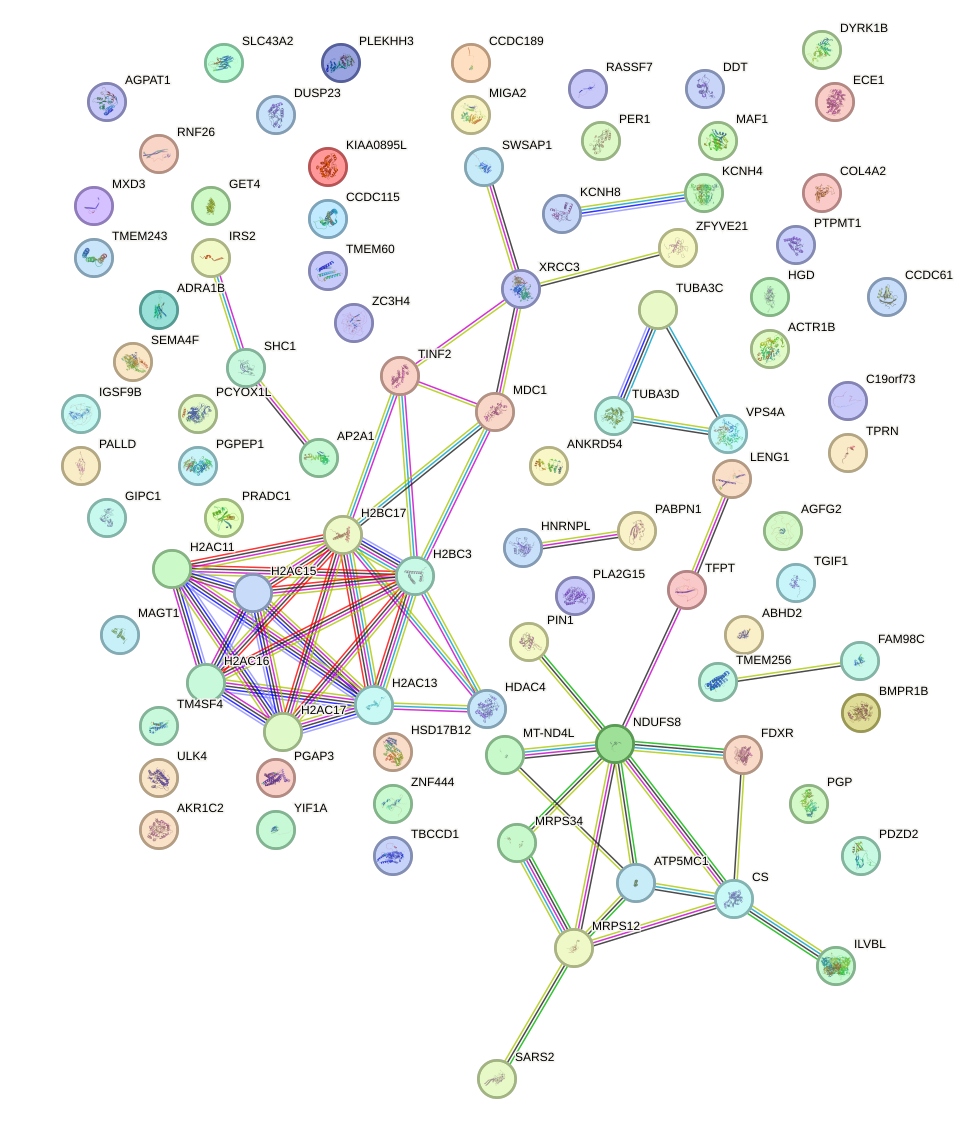
Network of associatons between targets according to the STRING database.
First level regulatory network of VSX1
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.1 | 1.1 | GO:0071394 | cellular response to testosterone stimulus(GO:0071394) |
| 0.1 | 0.2 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.1 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.0 | 0.2 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.0 | 0.1 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.1 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.0 | 0.2 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.0 | 0.1 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.0 | 0.1 | GO:0071139 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 0.0 | 0.2 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.3 | GO:0010836 | negative regulation of protein ADP-ribosylation(GO:0010836) |
| 0.0 | 0.3 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:0001994 | norepinephrine-epinephrine vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001994) |
| 0.0 | 0.1 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.0 | 0.1 | GO:0006114 | glycerol biosynthetic process(GO:0006114) |
| 0.0 | 0.2 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.2 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.1 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.3 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.2 | GO:0071395 | response to jasmonic acid(GO:0009753) cellular response to jasmonic acid stimulus(GO:0071395) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.2 | GO:0010748 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.0 | 0.2 | GO:0061051 | positive regulation of cell growth involved in cardiac muscle cell development(GO:0061051) |
| 0.0 | 0.2 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 0.1 | GO:0042246 | tissue regeneration(GO:0042246) |
| 0.0 | 0.1 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.0 | 0.1 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.0 | 0.0 | GO:0006258 | UDP-glucose catabolic process(GO:0006258) |
| 0.0 | 0.1 | GO:0010816 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.0 | 0.0 | GO:0070434 | positive regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070426) positive regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070434) |
| 0.0 | 0.1 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 | 0.0 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.2 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.3 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.0 | GO:0071106 | coenzyme A transport(GO:0015880) coenzyme A transmembrane transport(GO:0035349) adenosine 3',5'-bisphosphate transmembrane transport(GO:0071106) AMP transport(GO:0080121) |
| 0.0 | 0.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.0 | 0.3 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.1 | GO:0097196 | Shu complex(GO:0097196) |
| 0.0 | 0.2 | GO:0070435 | Shc-EGFR complex(GO:0070435) |
| 0.0 | 0.3 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.2 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.3 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.3 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.1 | GO:1990031 | pinceau fiber(GO:1990031) |
| 0.0 | 0.1 | GO:0031302 | intrinsic component of endosome membrane(GO:0031302) |
| 0.0 | 0.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.1 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0015039 | ferredoxin-NADP+ reductase activity(GO:0004324) NADPH-adrenodoxin reductase activity(GO:0015039) oxidoreductase activity, acting on iron-sulfur proteins as donors(GO:0016730) oxidoreductase activity, acting on iron-sulfur proteins as donors, NAD or NADP as acceptor(GO:0016731) |
| 0.1 | 0.2 | GO:0000994 | RNA polymerase III core binding(GO:0000994) |
| 0.1 | 0.3 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.1 | GO:0033981 | D-dopachrome decarboxylase activity(GO:0033981) |
| 0.0 | 0.1 | GO:0036440 | citrate (Si)-synthase activity(GO:0004108) citrate synthase activity(GO:0036440) |
| 0.0 | 0.2 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.0 | 0.2 | GO:0050815 | phosphoserine binding(GO:0050815) phosphothreonine binding(GO:0050816) |
| 0.0 | 0.4 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.1 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.1 | GO:0098519 | nucleotide phosphatase activity, acting on free nucleotides(GO:0098519) |
| 0.0 | 0.2 | GO:0018636 | phenanthrene 9,10-monooxygenase activity(GO:0018636) ketosteroid monooxygenase activity(GO:0047086) |
| 0.0 | 0.2 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.1 | GO:0000285 | 1-phosphatidylinositol-3-phosphate 5-kinase activity(GO:0000285) |
| 0.0 | 0.1 | GO:0004936 | alpha-adrenergic receptor activity(GO:0004936) |
| 0.0 | 0.3 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.2 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.1 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 0.0 | GO:0080122 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.4 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.3 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |